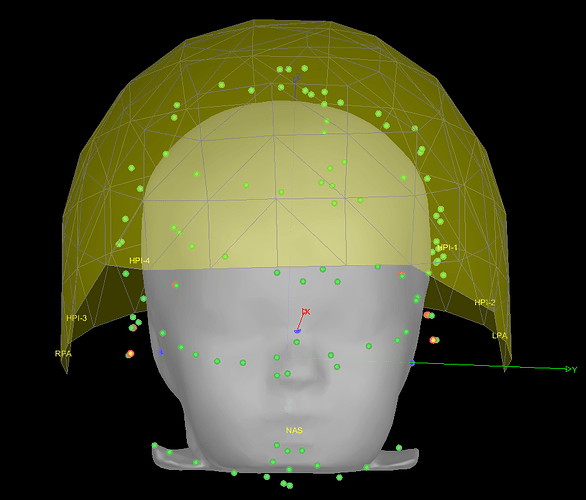
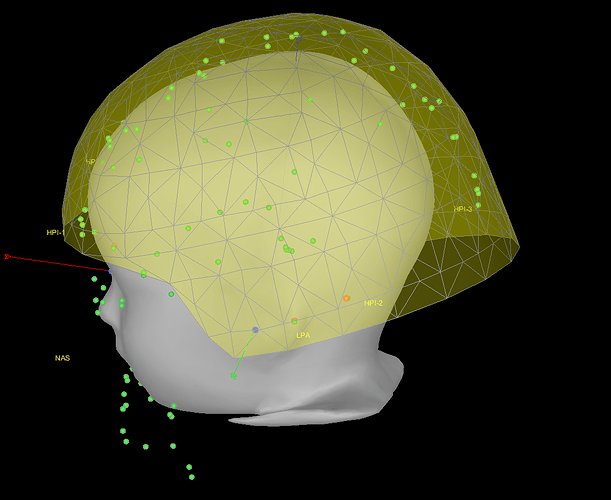
Yes, it is certainly better like that.
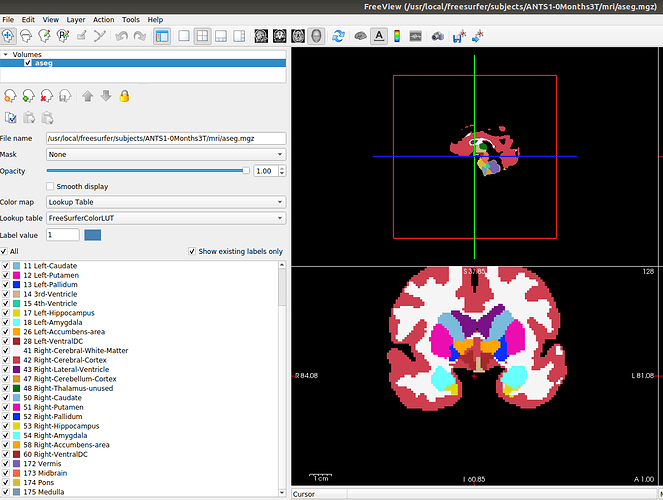
For reference, regarding the ASEG missing region issues, does Brainstorm use something else than the aparc+aseg.mgz file to determine the available regions when importing FreeSurfer folders? In the new templates, the aparc+aseg.mgz files contain the following regions for all templates:
['Left-Cerebral-White-Matter',
'Left-Cerebral-Cortex',
'Left-Lateral-Ventricle',
'Left-Cerebellum-Cortex',
'Left-Thalamus',
'Left-Caudate',
'Left-Putamen',
'Left-Pallidum',
'3rd-Ventricle',
'4th-Ventricle',
'Left-Hippocampus',
'Left-Amygdala',
'Left-Accumbens-area',
'Left-VentralDC',
'Right-Cerebral-White-Matter',
'Right-Cerebral-Cortex',
'Right-Lateral-Ventricle',
'Right-Cerebellum-Cortex',
'Right-Thalamus',
'Right-Caudate',
'Right-Putamen',
'Right-Pallidum',
'Right-Hippocampus',
'Right-Amygdala',
'Right-Accumbens-area',
'Right-VentralDC',
'Vermis',
'Midbrain',
'Pons',
'Medulla']
The following regions are available in all templates except the 0.5, 1, 2, 3 months templates:
['Left-Cerebellum-White-Matter',
'Right-Cerebellum-White-Matter']
The following regions are available in all templates except the 0.5, 1, 2, 3, 4.5, 18, and 24 months templates:
['Optic-Chiasm']
So the issue you previously observed should be resolved. If you still observe missing regions (except those mentioned here) when you'll process the updated templates, please let me know.
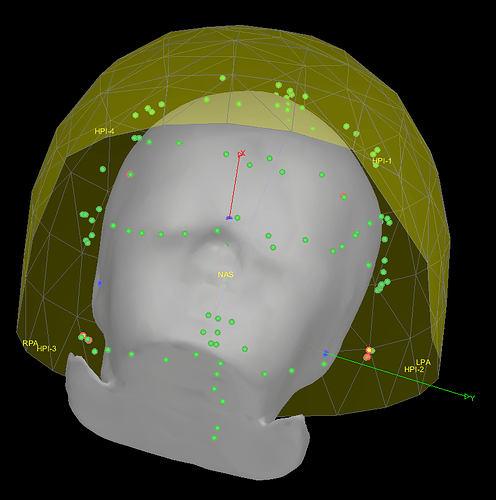
To your question "Do you have the description of all the 146 points (or at least of the 17 first)?", the 17 first points where actually duplicating the Nasion, LPA, and RPA. This has been removed, leaving 14 points. These 14 points are:
- left preauricular point (LPA)
- nasion (Nz)
- right preauricular point (RPA)
- anterior commissure (AC)
- posterior commissure (PC)
- inion (Iz)
- left mastoid (LMA)
- right mastoid (RMA)
- left outer canthii for EOG (LEOG)
- right outer canthii for EOG (REOG)
- left EMG cheek (LEMG)
- right EMG check (REMG)
- mid EOG on nose
- vertex (Cz)
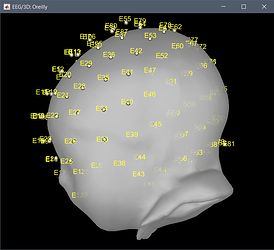
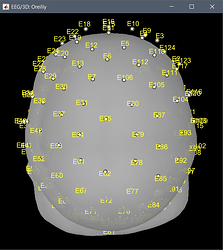
The other channels, are all EEG, numbered as you would expect for the corresponding montage.
You were also mentioning some inaccuracy for the position of the channels. These have now been projected to the closest point between their original position and the skin mesh.
Last time, I created the Brainstorm templates through the GUI. To improve reliability of this process, I will try to script it this time. Once I figured this out, the new templates should be ready and they should address the different concerns you had so far. Please let me know if I forget to address something.