Hi, all
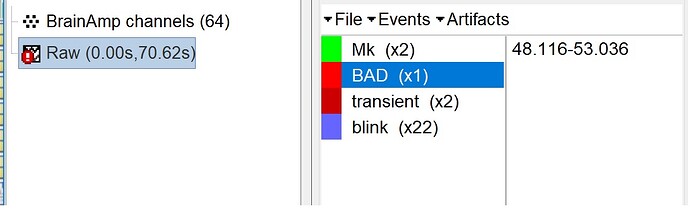
I found some segments in my EEG that I want to remove them from my analysis.
First of all, I've selected the bad segment with reject time segment but I don't know how can I remove this part from my signal.
In EEGLab when you are not interested in some segments with reject button you can remove it.
How can I do that in Brainstorm?
And another problem is when I want to import this data in database I can't do more process.
Best Regards,
Hamed
Hi Hamed,
When you import your data, the bad segments from your raw data will not be included (or rather, the corresponding epoch(s) will be marked as bad and not used in further processes). More info on bad segments can be found here: http://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#Bad_segments
For information on how to import your data once it is cleaned, refer to the following tutorial: http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching
More processes will be available once your data is imported and the relevant objects are dragged in the process bar at the bottom.
Cheers,
Martin
Thanks a lot Martin For your reply
Cheers,
Hamed
Hi Martin!
Thank you for your answer!
I understood that bad segments are not used in the following Brainstorm processing, but I’m still interested whether I can delete this bad segments forever in order to use the data in Matlab script. For example, if I run the File —> Export to Matlab command, I export all the data, including the bad segments, and then I need to delete this segments manually using the Events field in the data structure.
Can I do it somehow through the brainstorm?
Thank you!
Aleksandra
I don’t think so. @Francois?
Hi Alexandra,
No, there is no possibility to cut out the bad segments.
What would be possible is to import all the good segments, then export them. Otherwise, you import the entire file in the Brainstorm database, then remove the segments you don’t want in your own scripts. The data and events structures are documented in the sections “On the hard drive” of the introduction tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#On_the_hard_drive
https://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#On_the_hard_drive
Francois
Hi there Francois and Martin. I just want to make sure I understood correctly:
Suppose I have a continuous signal including three blocks and breaks between them. Breaks obviously contain a lot of heavy noise due to movement and talking, which I want to get rid of before running SSPs/ICAs as a way to optimise the results of such procedures. If I mark these long segments as bad, will it have the same effect as cutting the breaks out and concatenating the signal corresponding to my blocks (namely, SSPs/ICAs will "ignore" these segments of gross noise) or is it something I should do in Matlab before importing my raw file?
I hope I made myself clear with the wording. Thanks in advance.
1 Like
This is correct. Mark the breaks as bad segments and they will be ignored from the ICA and SSP computations.
To double-check the bad segments are excluded: mark as bad also the rest of the recordings, so that the entire file is marked with overlapping bad segments. Run the SSP or ICA process, it should return an error saying that no good data could be read from the input file.
1 Like
Dear Francois,
Let me know if I am right or not.
As you said the segments that are marked as bad segments,are excluded from the analysis.
Is that right about the source reconstructions?
First I marked bad segments, then I run ICA to remove the blink and noisy sources from my EEG data.
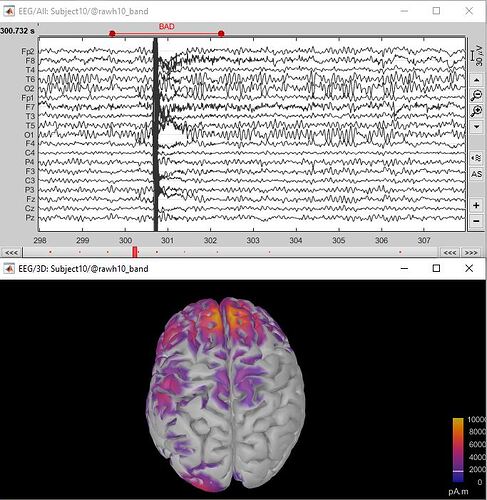
When I do source reconstruction, it seems that the bad segments are not excluded.
If you import this segment of data to the database, it would be marked as bad in the database.
It is excluded from the PSD computation and some other processes.
But other than that, the bad segment stays in your data, and if you ask Brainstorm to display the sources estimated in the middle of the artifact, it would show it.
You should not worry about this: the minimum norm estimates produces an inverse solution that is independent on the data. It produces a linear inverse operator which depends only on the forward model and the noise covariance matrix. The recordings themselves do not impact the source estimation, so it does not matter whether you have bad segments or not in your recordings...
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation
Thank you for your response.
I appreciate it.
Sima.