Dear Raymundo
Thank you very much for all your advice.
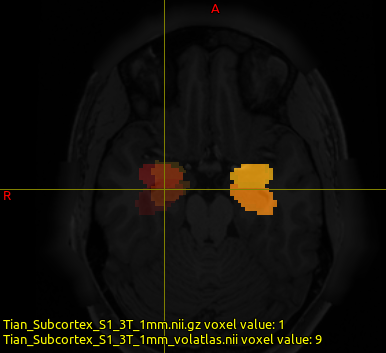
I have written a script, and the resulting output looks good. Could you please take a look at my script to verify that everything is correct? In this script, I iterate over three subjects and use the Tian Subcortex parcellation in MNI space (Tian_Subcortex_S1_3T_1mm.nii.gz).
% Script generated by Brainstorm (26-Sep-2024)
% Input files
sFiles = [];
SubjectNames = {'100206','100307','100408'};
RawFiles = {...
'/home/cris/Escritorio/ProbarPipelineBrainstorm/100206/T1w_acpc_dc_restore.nii.gz', ...
'/home/cris/Escritorio/ProbarPipelineBrainstorm/100307/T1w_acpc_dc_restore.nii.gz', ...
'/home/cris/Escritorio/ProbarPipelineBrainstorm/100408/T1w_acpc_dc_restore.nii.gz', ...
'/home/cris/Escritorio/ProbarPipelineBrainstorm/Tian2020MSA/3T/Subcortex-Only/Tian_Subcortex_S1_3T_1mm.nii.gz'};
% Start a new report
bst_report('Start', sFiles);
for iSubject = 1:3
% Process: Import MRI
sFiles = bst_process('CallProcess', 'process_import_mri', sFiles, [], ...
'subjectname', SubjectNames{iSubject}, ...
'mrifile', {RawFiles{iSubject}, 'ALL'}, ...
'nas', [0, 0, 0], ...
'lpa', [0, 0, 0], ...
'rpa', [0, 0, 0], ...
'ac', [0, 0, 0], ...
'pc', [0, 0, 0], ...
'ih', [0, 0, 0]);
% Process: MNI normalization
sFiles = bst_process('CallProcess', 'process_mni_normalize', sFiles, [], ...
'subjectname', SubjectNames{iSubject}, ...
'method', 'maff8', ... % maff8:Affine registration using SPM mutual information algorithm.Estimates a simple 4x4 linear transformation to the MNI space.Included in Brainstorm.
'uset2', 0);
% Process: Import MRI
sFiles = bst_process('CallProcess', 'process_import_mri', sFiles, [], ...
'subjectname', SubjectNames{iSubject}, ...
'mrifile', {RawFiles{4}, 'ALL-MNI-ATLAS'}, ...
'nas', [0, 0, 0], ...
'lpa', [0, 0, 0], ...
'rpa', [0, 0, 0], ...
'ac', [0, 0, 0], ...
'pc', [0, 0, 0], ...
'ih', [0, 0, 0]);
% Get Subject structure and their MRIs
sSubject = bst_get('Subject', SubjectNames{iSubject});
% Find the parcellation MRI filename in the array sSubject.Anatomy
iMniAtlas = find(contains({sSubject.Anatomy.Comment}, 'Tian_Subcortex_S1_3T_1mm (MNI-linear)', 'IgnoreCase',true));
% Save MNI atlas as NIfTI file 'new_MRI.nii'
export_mri(sSubject.Anatomy(iMniAtlas).FileName, strcat('/home/cris/Escritorio/ProbarPipelineBrainstorm/',SubjectNames{iSubject},'/Tian_Subcortex_S1_3T_1mm_volatlas.nii') );
end
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
% bst_report('Email', ReportFile, username, to, subject, isFullReport);
% Delete temporary files
% gui_brainstorm('EmptyTempFolder');
Best regards
Cristóbal