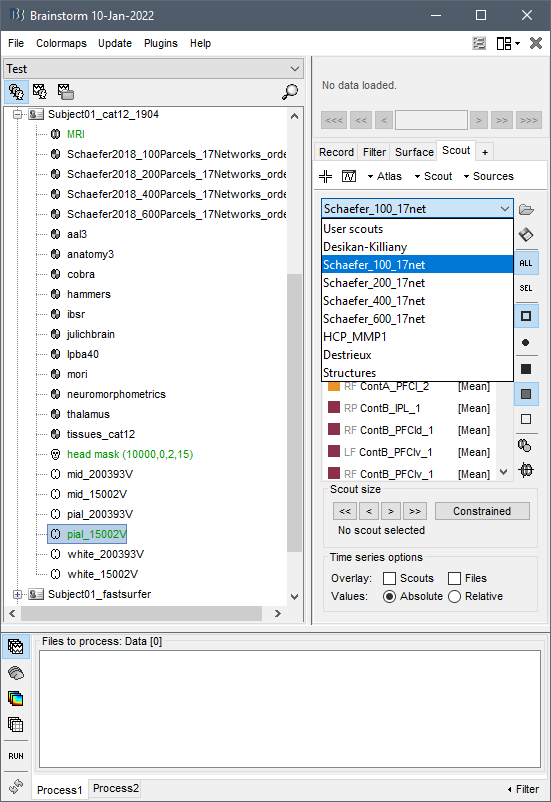
Can I use the segmentation of CAT12 in the default anatomy folder (as I don't have individual MRI) or do I need to do each subject manually?
You can use the default MNI ICBM152 as it is, without any modification (FreeSurfer segmentation).
As I said before, if you need the Shaeffer segmentation, run CAT12.
When I try the segmentation with the MNI152 in the subject folder, it works, but not in the default folder.
Do you mean that if you right-click on the file Default anatomy / MRI: ICBM152 > MRI segmentation > CAT12, you get an error?
If so, please copy-paste the full error message you get, together with a screen capture of what your database looks like.
I cannot compute my source if I first segment the MNI152 (the BEM head model doesn't work), but I can do the reverse (compute source and then segment). Is it the usual way ?
If you run CAT12 on the ICBM152 brain, you would need to recompute the BEM surfaces after:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem
If you get an error with OpenMEEG computation: check the workarounds in the tutorial, then if nothing works please report the full error message in a different thread.
At the end, I can apply the Schaefer atlas on my source reconstuction,
There is no need to "apply" anything.
The Schaefer atlas is readily available on the cortex surfaces when running the MRI segmentation with CAT12.
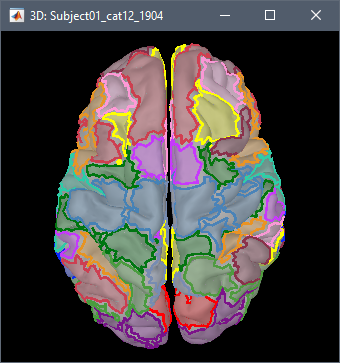
I can see the scoots related to the selected atlas. But there are values with a scale (similar to the one in source reconstruction). I am not sure to understand what it is.
I'm not sure what you are referring to.
Please post screen captures and explain better your question.