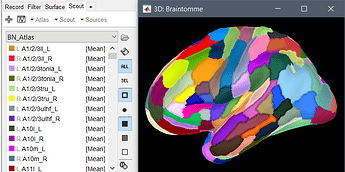
Thanks for the files.
I realized that this is a parcellation scheme that you can project to any subject you process with FreeSurfer, following the instructions in the Brainnetome Atlas in Freesurfer and Workbench.
I've just added the instructions to load the *h.BN_Atlas.annot atlases when importing the FreeSurfer anatomy. Now whenever these files are available in the FreeSurfer folder "label", they'll be imported automatically by Brainstorm.
Import FreeSurfer: Added Braintomme atlas · brainstorm-tools/brainstorm3@d4b8e04 · GitHub
- Update Brainstorm
- Project the Braintomme atlas to your subject folder: NITRC: Brainnetome Atlas Viewer: File Release Notes and Changelog
- Import the FreeSurfer folder: https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer