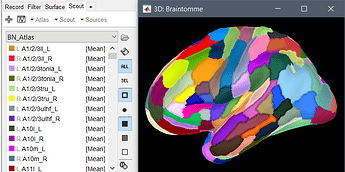
Hi everyone, I need a suggestion. I'm trying to import brainnetome atlas in the brainstorm default anatomy surface with 15002 vertices but I think I'm doing something wrong. This is what I did:
I downloaded the .annot brainnetome label and surface, in the folder I found the two hemispheres separate.
I imported the surface of each hemisphere separately, importing the atlas label for each hemisphere.
Then, I merged the two hemispheres obtaining a surface with 123000 vertices with the complete brainnetome atlas. I downsampled the surface to 15002 and the I exported from brainstorm the .mat atlas.
I tried to import the brainstorm exported .mat atlas in the default anatomy surface, although even if I did not receive an error message when I plot the surface I obtain several colored dots on the surface instead of the atlas.
I'm sure that it is not correct to import an atlas obtained from a surface to another, but considering that brainnetome atlas is available on the freesurfer surface, what could I do to import it also on the brainstorm default anatomy?
Thanks!
There is no easy solution for this. The surfaces created by FreeSurfer are not exchangeable between subjects, the vertices do not match point-to-point between surfaces, they may even have different numbers of vertices. Therefore it is not possible to use the .annot files (= a texture that associates each vertex of a surface to a label) computed on one subject for another subject.
Do you have access to a full FreeSurfer folder (including T1.mgz, ASEG*.mgz, lh.pial, etc.) for the Braintome atlas?
We could import this into Brainstorm and make it a new template.
Yes I do. If you want I could share it with you and/or Martin. Let me know.
Thanks
Yes please. You could zip the entire FreeSurfer folder, upload it anywhere and post the download link here.
Thanks
This is the link:
https://mega.nz/#!QkBG2AjI!MnZTo0BB6HrFo02A3jPsmjp3o3GKE0BhW6j0JSIFEJA
Let me know if you have any problem in downloading the folder
Thanks for the files.
I realized that this is a parcellation scheme that you can project to any subject you process with FreeSurfer, following the instructions in the Brainnetome Atlas in Freesurfer and Workbench.
I've just added the instructions to load the *h.BN_Atlas.annot atlases when importing the FreeSurfer anatomy. Now whenever these files are available in the FreeSurfer folder "label", they'll be imported automatically by Brainstorm.
Import FreeSurfer: Added Braintomme atlas · brainstorm-tools/brainstorm3@d4b8e04 · GitHub
- Update Brainstorm
- Project the Braintomme atlas to your subject folder: NITRC: Brainnetome Atlas Viewer: File Release Notes and Changelog
- Import the FreeSurfer folder: https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer
That sounds awesome! Does it mean that in case I use the default anatomy if I want to use the atlas I just need to use FSaverage instead of the ICBM 152, right?
Thanks!
I've just uploaded a new version of the FSAverage atlas, which includes the Brainetomme atlas:
https://github.com/brainstorm-tools/brainstorm3/commit/ffbdb4dd8ef6207aa58d425929f047ede96d90e9
To get it: update Brainstorm again, then right-click on an subject folder in the anatomy view > Use template > Download: FSAverage_2020.
If you have the MRI of your subjects, you should rather run FreeSurfer and project the Braintomme atlas on their own cortical surfaces.
Hi Francois,
I tried "right-click on an subject folder in the anatomy view > Use template > Download: FSAverage_2020"
and got:
** Error: Impossible to download template:
** Error: File to download does not exist.
Is this still available?
I've just tested it, and it seems to work.
Make sure you don't have any network restriction that prevents you from downloading: http://neuroimage.usc.edu/bst/getupdate.php?t=FSAverage_2020
Thanks Francois, it seems I have additional network restrictions that must have been added recently as I am now prevented from downloading other atlases which which I able to do without issue previously.
You can download the templates you need from the Brainstorm download page.
Then copy the .zip files manually to the folder: $HOME/.brainstorm/defaults/anatomy
(do not unzip them)
Thanks, I had actually done this before you responded but I did unzip the file and it seemed I was able to see and load the default anatomy within Brainstorm. I saw the ICBM152 folder was unzipped so just tried to copy the format.
Why is it preferable not to unzip? I will replace with the zipped version regardless.
Both work.
Just do not add it to the brainstorm3 folder, but to the .brainstorm folder, so that it doesn't disappear when you update Brainstorm.
Excellent. Thank you.
Hi Francois,
Posting in this thread as it follows on from my network restriction issue. If I have the MRI viewer open and I click the "Click here to compute MRI transformation" link, I get the popup which says that displaying MNI coordinates requires the download of additional atlases, which doesn't work for me given the network restrictions. Can please let me know which atlases are required so that I can download them manually?
You need to download: http://neuroimage.usc.edu/bst/getupdate.php?t=SPM_TPM
And unzip it as: $HOME/.brainstorm/defaults/spm/TPM.nii
If you can connect temporarily your computer to a different network, you can simply run brainstorm workshop and it will download what it needs for offline analyses:
Hello, Francois
I have individual MRI scans for each subject (I used freesurfer before)
I projected my data (source activity) on the default anatomy and then did as you suggest: right-click on a subject folder (in my case is Default anatomy) in the anatomy view > Use template > Download: FSAverage_2020.
Despite I got scouts of Brainetomme atlas, the source activity itself began to look like colored dots on the cortex surface
Is there a way to get scouts from Braintomme atlas and have normal source activity but not running freesurfer again?
Thanks
I projected my data (source activity) on the default anatomy and then did as you suggest: right-click on a subject folder (in my case is Default anatomy) in the anatomy view > Use template > Download: FSAverage_2020.
If you want to use the FsAverage as the default anatomy in your protocol, you need to do this BEFORE projecting your individual source results on the default anatomy. What you did here deleted the anatomy support of your source projections on the template.
The best procedure is definitely what is discussed above in this thread: use FreeSurfer to project the Brainnetome atlas to the subject space and then import the subject anatomy in your Brainstorm database. And then work at the ROI level in the subject space and never rely on the projection of the full source maps to the template space.
https://www.nitrc.org/frs/shownotes.php?release_id=3459