Hi,
Brainstrom has a template called ICBM152, available for download.
http://neuroimage.usc.edu/bst/download.php
Could you please send me the .nii format of MR image of this template? ( I can only export it to matlab with .mat format, but I need .nii one)
I guess it is a little bit modified version of this one http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin6
and also, is it possible to use the method mentioned in this page
http://neuroimage.usc.edu/forums/archive/index.php/t-1672.html
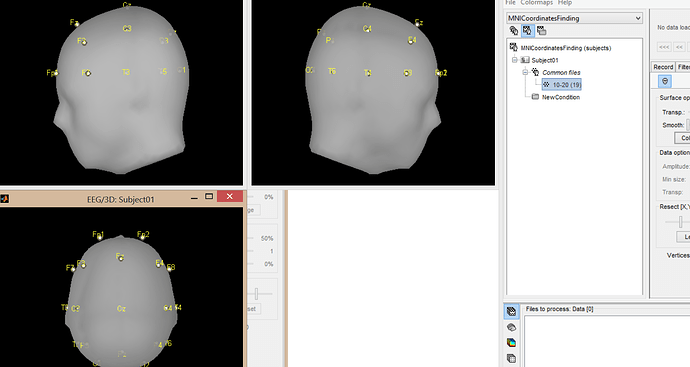
- Create a new subject, using the default anatomy
- Create a condition, right-click on it > Use default EEG cap > Colin27 > 10-20 (19)
- Right-click on the channel file > Export to Matlab > ChannelMat
- Right-click on the MRI file > File > Export to Matlab > sMri
- Electrodes locations of channel #i in SCS:
xyz_scs = ChannelMat.Channel(i).Loc’;
- Convert to MRI coordinates (millimeters):
xyz_mri = cs_scs2mri(sMri, xyz_scs’ .* 1000)’;
- Convert to MNI coordinates (meters):
xyz_mni = cs_mri2mni(sMri, xyz_mri’)’ ./ 1000;
to get the coordinate of EEG locations exactly on this ICBM152?
Thank you so much!
Hi,
I used above method to find EEG coordinates for ICBM152 template.
and created a new subject in brainstorm with individual anatomy and changed it to ICBM152 template.
and used xyz_scs variable as the channel locations for this subject.
0.106535168779891 0.0334649524249551 0.0451641266156433
0.106640217990819 -0.0330536117360768 0.0451641266156433
0.0635420658319772 -0.0579336973923552 0.0944613868782839
0.0627452329249089 0.0589681965916031 0.0931672141545323
-0.00395420690389706 0.0676932150300701 0.112105016730153
-0.00338874956032856 -0.0699441200296035 0.111539559386584
-0.0579482443157264 -0.0550570861801225 0.0943121392483022
-0.0593197611025517 0.0541566513477002 0.0931672141545323
-0.0880220096988024 -0.0357966453097275 0.0485929185827068
-0.0861273094263320 0.0348364692117805 0.0485929185827068
0.0571151862015011 -0.0748754440274123 0.0448155928455012
0.0567786044493769 0.0745899589421239 0.0451252480574554
-0.00282329221676006 -0.0846345716872656 0.0448155928455012
-0.00299831472786463 0.0833919711242061 0.0456637788608540
-0.0557874331544380 -0.0693988065869499 0.0472214017958814
-0.0572624859223136 0.0669584323604577 0.0465356434024687
-0.0716634121839804 -0.00150872563909259 0.106519423991099
0.0737173672195119 0.000548549541145540 0.121296217722143
-0.00461390713806037 -0.000683329574176161 0.147161451122660
As you see they are not fitted on the skull:
I thought maybe I should have used xyz_mni, but I tried that one too and it was very much worse!
Hello,
The standard electrode positions you use are aligned on the Colin27 brain, not on the ICBM152. I will do that at some point, but it is not available now.
If you want electrodes aligned on the skin surface, use the Colin27 template.
To export an MRI as .nii from Brainstorm: right-click on it > File > Export to file > Select the file format “NIfTI”.
Cheers,
Francois
Hi Francois,
Thank you very much, I got the .nii format!
Actually for our case we need to use ICBM152.
I projected the Colin27 electrode locations on ICBM152 template just for now, and used them for registration. But whenever you have the locations on ICBM152, please let me know.
Best regards,
Haleh
The two templates Colin27 and ICBM152 share the same head surface, so you should be able to just click on the button “Refine registration using head points”.
I will fix the current ICBM152 model so that it uses the same NAS/LPA/RPA points as the Colin27 brain.
Thank you for bringing up this issue.
Oh, Thats great!
Thanks for your consideration!
Best,
Haleh