Hello Nirstorm team
First, thank-you for the amazing program!
I am trying to generate sensitivity models for my montage with subject-specific anatomy following this tutorial: Compute head model from fluence · Nirstorm/nirstorm Wiki · GitHub.
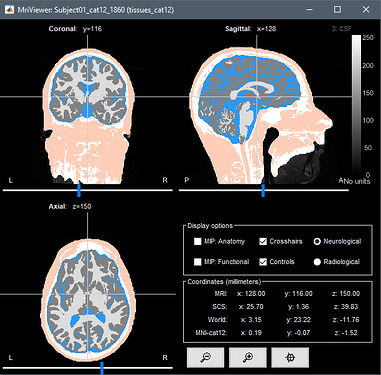
So far I have been able to load in the specific anatomy for several participants and create the segmentation_5tissues volume using Fieldtrip for each of them. I then needed to recode the tissues to be 1:scalp, 2:skull, 3:CSF, 4:gray, 5:white matter which I did in Matlab by editing the Labels variable in the segmentation_5tissues .mat file
(CAT12 - 5 tissue segmentation). The fNIRS montage also appears to be loaded correctly.
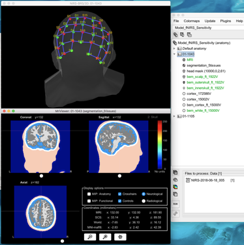
Then I attempted to calculate the fluences from the optode positions using the individual subject anatomy via the ‘compute fluences for optodes by MCXLab’ function.
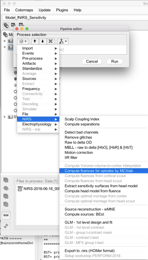
I get the following error (Undefined function 'surfacenorm' for input arguments of type 'double'). I believe I have MCXLab installed correctly since I can type “help mcxlab” at the command line and get some help documentation.
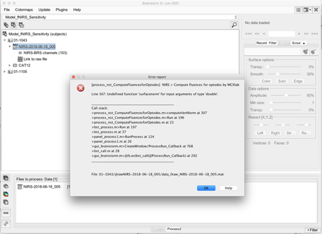
Is there additional information/instructions you have that I can follow? There are a few TODO entries in the tutorial above, possibly there are additional documents available now?
Thanks very much
Helen.
Hello @hcarlson,
Thanks a lot for your interest in nirstorm.
This is a known issue: this process is using iso2mesh to compute the normal vector to the head mesh.
Can you make sure you are using the latest version of NIRSTORM?
Otherwise, you can use the plugin manager system to download and load iso2mesh before computing the fluences.
Also note that in the latest version of Nirstorm, you no longuer have to change the segmentation value; we added an option in the process to chose how is your MRI segmented.
Btw, note that when doing this operation you should not only change the label but also the value themselves. This can be donne by doiing something like:
tmp2 = tmp; % Where tmp is your segmentation file
tmp2.Cube ( tmp == 1 ) = 5;
Regards,
Edouard
Hello all
Thanks very much for your quick replies, I have updated my Brainstorm and Nirstorm to the latest versions and have loaded iso2mesh via the plugin manager.
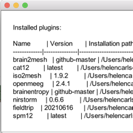
I then realised that FieldTrip made some errors in the segmentation process (my patient brains have strokes that need to be classified as CSF) and that CAT12 does a much better job segmenting the lesioned areas. So I have managed to use expert mode in CAT12 (externally via the CAT12 GUI) to output the additional tissue types needed. Once loaded into Nirstorm, via the “Import anatomy folder” option, only three tissue types appear in the tissues volume despite the presence of the other tissue types in the same directory (and they are flipped left-right).
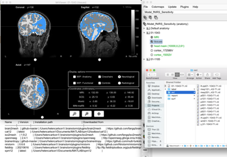
If I run the CAT12 segmentation from within Nirstorm via the "MRI Segmentation/CAT12" option then again, only 3 tissues are included in the tissues volume (but the flipping problem goes away). Is there a way to load the rest of the tissue types from CAT12 required for the fluence calculations?
I’m also getting a new error when I attempt to compute fluences, likely because the tissue types loaded are incorrect.
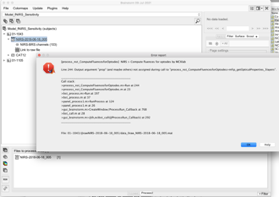
Thank-you
Helen
Hello all
I have come up with a different way to accomplish my goal (calculating fNIRS sensitivity with individual anatomy) and wanted to ask you whether it is reasonable to estimate sensitivities in this way.
I have loaded in the Colin27_4NIRS atlas into Nirstorm as well as a datafile using my fNIRS montage. I then calculated the sensitivity profile for this montage using the pre-calculated fluences for the Colin27 atlas.
I then was able to right click on the summed sensitivities volume for Colin27 and select Project sources/Other subjects and I selected a patient’s brain. The resulting sensitivity profile looks reasonable and my question is, is this a valid way to estimate the sensitivity profile for my fNIRS montage?
Colin27:
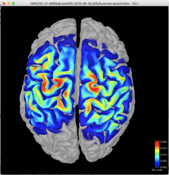
Patient:
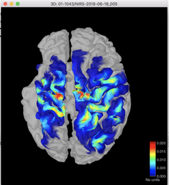
Thank-you for your help
Helen
Hello Helen,
Sorry for the delay in my reply;
No I don't think this is a good idea since the sensitivity map (eg the propagation of the light) is highly dependent on the subject anatomy. (Especially, CSF is influencing a lot the propagation of light). I don't think doing a simple projection from sensitivity on colin27 will work well.
We should instead try to figure out why the segmentation is not working. I guess it won't be possible for you to share with me the MRI and localisation of the nirs ?
Can you provide more detail on how you run cat12 ? @Francois; could you help us on that since it's might be more an issue with Brainstorm ? As a reminder, for nirs, we need to create a segmentation file with the 5 tissue (scalp; skull; CSF, GM, WM)
Regards,
Edouard
Hi Edouard
Thank-you for your reply. I can share the deidentified MRI and NIRS files with you offline if I can email you a link directly. That may be the easiest way to proceed.
For the segmentations in CAT12 I did it two different ways attempting to solve the issue:
Method 1. Segment in Brainstorm by loading the raw T1-weighted anatomical file and then selecting MRI segmentation/CAT12. This runs CAT12 and correctly loads three tissue types (GM, WM, CSF) though I suspect the errors I was getting when trying to calculate the fluences from optode (original post) was because there are only three tissue types rather than five.
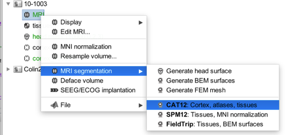
Method 2. Run via CAT12 in expert mode by running CAT12 directly via Matlab, selecting Expert mode, then selecting in the batch editor Grey matter/native space and White matter/native space “Yes”.
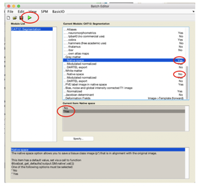
This gives me six tissue type volumes but when I load the folder (using “import anatomy folder”/CAT12 + thickness) into Brainstorm/Nirstorm, they are flipped around the left/right axis (i.e., the stroke is on the wrong side) and again only loads the first three tissue volumes.
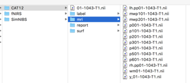
It may be that the automatic loading is loading the p001 file which only contains the first three tissue types but I'm not too sure why the resulting loaded tissue segmentations are reversed in the left-right axis.
If there is an easier way to model the NIRS sensitivity, I would be happy to try that.
Thanks very much
Helen
Hi;
You can send me the data at Edouard.delaire@concordia.ca; I should be able to have a look at it next Monday.
Edouard
Sorry for the response delay...
Method 1 . Segment in Brainstorm by loading the raw T1-weighted anatomical file and then selecting MRI segmentation/CAT12. This runs CAT12 and correctly loads three tissue types (GM, WM, CSF) though I suspect the errors I was getting when trying to calculate the fluences from optode (original post) was because there are only three tissue types rather than five.
When running CAT12 from Brainstorm, this gives you 5 tissue classes: gm, wm, csf, skull, skin:
https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12#Tissue_classification
An execution example with the latest CAT12 version:
If you obtain anything else than this:
@Francois; could you help us on that since it's might be more an issue with Brainstorm ? As a reminder, for nirs, we need to create a segmentation file with the 5 tissue (scalp; skull; CSF, GM, WM)
What kind of extra help do you need?