Hello!
I have been trying to calculate the WPLI at sensor level, without success. I already tried several combinations of parameters and nothing, always comes out some error:
BST> Error: There are 57000 abnormal NaN values in this file, check the computation process.
Oops select
or it tells me that the file time is too short for the resolution.
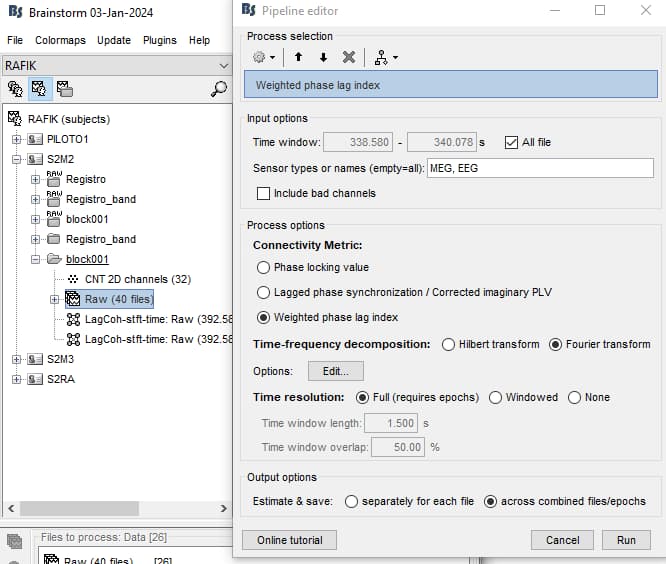
I use 1.5 seconds epochs, and Matlab default bands (Alpha, Beta, etc...).
Has anyone already managed to do it?
Which parameters should I use?
Reviewing articles I see that the original WPLI uses Fourier, even so I have already tried Hilbert and nothing.
Please help!!!
Thanks in advance!!!
pd: this only happens to me with WPLI, I've been able to use lagged coherence without problems.

Hi,
It runs fine for me on tutorial data, with Fourier and no time resolution. It's probably an issue with the specific options you're trying. Can you either describe all of them for the case that produces the NaN values, or show a screenshot?
Time resolution with Fourier can be confusing because there's 2 levels of windowing, which must be compatible. First, the data is windowed for time, then each of these "time segments" is windowed again for Fourier. It's fine if each has only 1 window if you have multiple epochs, e.g. 1s time windows and 1s Fourier windows. Some of the errors you're getting are probably due to those settings.
Hi
I tried once more:
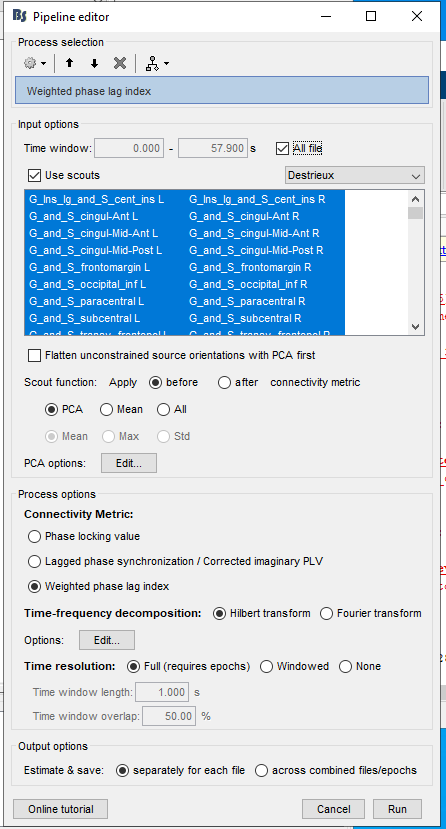
Epochs are 1.5 sec
I used these parameters:
and then:
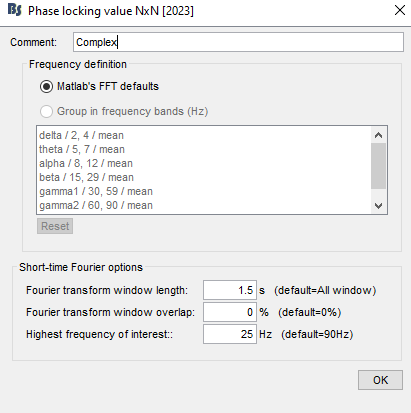
apparently works fine, but when plotting it, the following message is displayed:
BST> Error: There are 6120 abnormal NaN values in this file, check the computation process.
Oops select
435 Connectivity measure loaded
>>
note: it does make the graph, but if you check the console this error appears.
UPDATE: I tryed with another file (from other subject), this time I used a FFT window of 1 sec, and got the same error message 
Edit: I get the warning when opening the resulting files as well. Looking into it further.
It works fine for me even with "full resolution", when selecting 1s windowing (my epochs were 1s long). By the way, do you really want connectivity across MEG and EEG channels together?
By the way, "Matlab FFT defaults" returns each frequency separately. It's not "default bands" as you said, but basically "no bands". The box with band definition is greyed out in that case: it only applies when you select "group in frequency bands".
I tried it again, this time I let the box for Channel type (EEG and MEG) empty, but got the same message,
I share here the original Neuroscan file, maybe you can check it please:
Note: I marked the lasts 3 seconds of this file as Artifact (when processing it with BS)
I hope this helps
thanks in advance
if it's empty, it means all channels, so it's even worse. What you probably want is MEG only? MEG & EEG is possible, but not common.
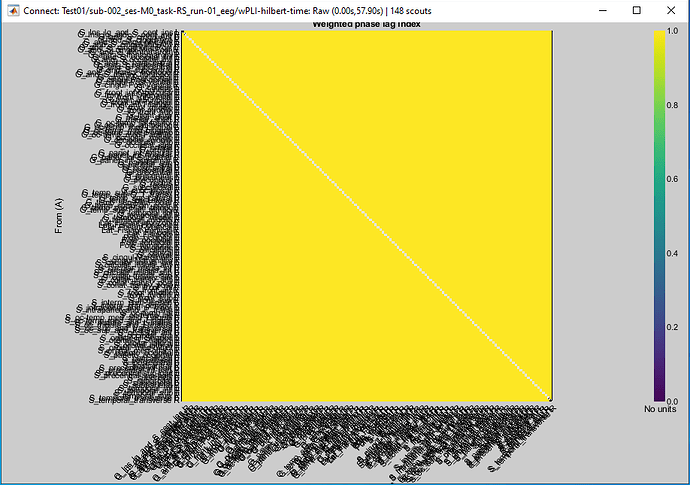
For me, the result is ok. The NaN values appear only on the diagonal, which I believe is fine. And the resulting connectivity matrix does load and display fine here. So this warning is not really an issue.
But I only get one line of warning about NaN values. I don't get "Oops select" or "X connectivity measures loaded". Are you running a "clean" (no custom modifications) and recent version of Brainstorm?
I see, it is not an error or problem then, is just the diagonal!
thank you very much for clarifying that!!

and yes, I'm using the last version of BS, with no modifications
Still not sure where the "Oops select" message comes from, but I found the 3rd message is when displaying the graph image. So yes I'd say there are no problems here.
We might want to change the message about NaN values to be less scary, @Raymundo.Cassani when you're back 
1 Like
Hello there,
We have the same issue with our data (source EEG). This is new and we did not have any issue to compute wPLI data last year (the process has more option now since the latest brainstorm update).
This is the case with the Hilbert and the PCA option (regardless of the PCA option selected).
This is not the same issue, as we determined there was no issue for the NaN message. 
It's normal to get 1 everywhere if you compute/save on single files.
@Raymundo.Cassani we might want to prevent/give error for "save separately for each file" when "full" time resolution is selected. This combination of options is not possible.
So you cannot compute PLI on a single recording ?
The message remains when unchecking the "full time resolution" box
The message is not an error, it's because the diagonal elements are NaN, which is fine.
WPLI on a single point always gives 1. The formula require averaging. So you can't do full time resolution AND save each file separately. For full time resolution, you need to average across epochs.
![]()