Hello everyone
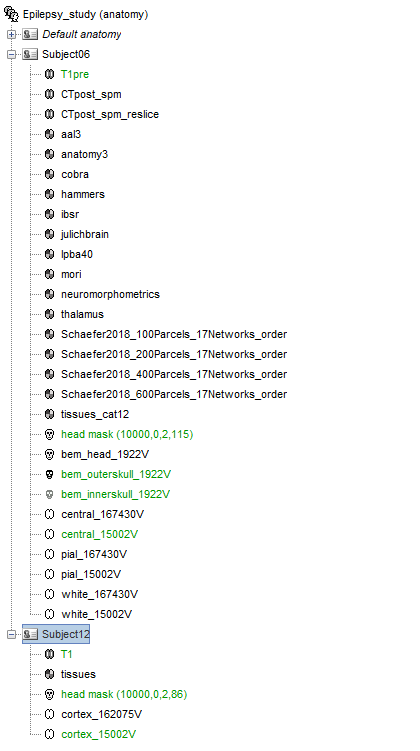
I am very new to Brainstorm and trying to figure out how to do group-level analyses of intracranial ERP data. From the tutorials online I have understood that the first proces is segmentation of the MRI on an individual level, and I have opted to use the software CAT. For the first subject this went well, however with the second subject (sub12) I can see that not everyting is loaded into Brainstorm (as was the case for the first subject, i.e. sub06).
Any ideas what I am doing wrong?
Many thanks!
It seems that the segmentation of subject 12 was not completed.
Can you run it and check the display errors/messages on the terminal?
Yes that is the thing I don't understand, MATLAB nor Brainstorm displayed any errors.
14-Aug-2023 16:03:58 - Running job #1
------------------------------------------------------------------------
14-Aug-2023 16:03:58 - Running 'CAT12: Segmentation'
Your version of CAT12 is up-to-date.
------------------------------------------------------------------------
CAT12.8.2 r2170: 1/1: .\.brainstorm\tmp\cat12_230814_160334\Subject12.n
------------------------------------------------------------------------
SANLM denoising (medium) 63s
Internal resampling (0.45x0.45x3.00mm > 1.00x1.00x1.00mm) 2s
APP: Rough bias correction
Initialize 5s
Estimate background 4s
Initial correction 5s
Refine background 3s
Final correction 4s
Final scaling 7s
32s
Correct center-of-mass 3s
Affine registration 14s
SPM preprocessing 1 (estimate 1 - TPM registration): 82s
SPM preprocessing 1 (estimate 2): 102s
SPM preprocessing 2 (write)
Write Segmentation 25s
Update Segmentation 20s
Update Skull-Stripping 37s
Update probability maps 7s
89s
Global intensity correction 14s
SANLM denoising after intensity normalization (medium) 12s
Fast Optimized Shooting registration 44s
Local adaptive segmentation (LASstr=0.50)
Prepare maps 5s
Prepare partitions 2s
Prepare segments (LASmod = 1.00) 14s
Estimate local tissue thresholds (WM) 15s
Estimate local tissue thresholds (GM) 18s
Intensity transformation 0s
SANLM denoising after LAS (medium) 21s
78s
ROI segmentation (partitioning)
Atlas -> subject space 10s
Major structures 4s
Ventricle detection 12s
Blood vessel detection 9s
WMH detection (WMHCstr=0.50 > WMHCstr'=0.12) 26s
Manual stroke lesion detection 0s
Closing of deep structures 2s
Side alignment 4s
Final corrections 3s
71s
Blood vessel correction (BVCstr=0.50) 1s
Amap using initial SPM12 segmentations (MRF filter strength 0.06) 62s
AMAP peaks: [CSF,GM,WM] = [0.35±0.06,0.68±0.08,0.98±0.05]
Final cleanup (gcutstr=0.25)
Level 1 cleanup (ROI estimation) 4s
Level 1 cleanup (brain masking) 3s
Level 2 cleanup (CSF correction) 1s
Level 3 cleanup (CSF/WM PVE) 2s
11s
Optimized Shooting registration with 2.50:-0.25:1.50 mm (regstr=0.50)
Template: "C:\Users\joerlema\.brainstorm\plugins\cat12\cat12\templates_MNI152NLin2009cAsym\Template_0_GS.nii"
1 | 2.50 | 0.0843 0.0000 0.0843 | 32.0000
2 | 2.50 | 0.0817 0.0011 0.0828 | 29.4886
3 | 2.50 | 0.0809 0.0017 0.0827 | 26.9772
4 | 2.50 | 0.0806 0.0019 0.0825 | 24.6107
5 | 2.50 | 0.0803 0.0021 0.0823 | 22.6548
6 | 2.50 | 0.0800 0.0022 0.0822 | 20.6989
7 | 2.50 | 0.0797 0.0023 0.0820 | 18.9688
8 | 2.50 | 0.0795 0.0024 0.0818 | 17.4455
9 | 2.50 | 0.0792 0.0024 0.0816 | 15.9223
10 | 2.50 | 0.0789 0.0025 0.0814 | 14.6627
11 | 2.50 | 0.0786 0.0026 0.0812 | 13.4764
12 | 2.50 | 0.0783 0.0027 0.0809 | 12.3015
13 | 2.50 | 0.0780 0.0028 0.0807 | 11.3776
14 | 2.50 | 0.0777 0.0028 0.0805 | 10.4537
15 | 2.25 | 0.0769 0.0029 0.0797 | 9.5920
16 | 2.25 | 0.0741 0.0043 0.0784 | 8.8725
29 | 2.00 | 0.0753 0.0019 0.0772 | 3.3283
30 | 2.00 | 0.0692 0.0044 0.0736 | 3.1221
31 | 2.00 | 0.0673 0.0054 0.0727 | 2.9160
43 | 1.75 | 0.0665 0.0030 0.0696 | 1.5785
44 | 1.75 | 0.0622 0.0054 0.0676 | 1.5194
45 | 1.75 | 0.0608 0.0062 0.0670 | 1.4626
57 | 1.50 | 0.0596 0.0045 0.0641 | 1.0900
58 | 1.50 | 0.0563 0.0067 0.0630 | 1.0730
59 | 1.50 | 0.0553 0.0073 0.0626 | 1.0579
Shooting registration with 2.50:-0.25:1.50 mm takes 196s
Prepare output 15s
211s
Jacobian determinant (RMS): 0.011 0.038 0.058 0.085 0.111 | 0.118914
Template Matching: 0.084 0.222 0.202 0.182 0.166 | 0.165932
Write result maps 168s
Surface and thickness estimation
lh:
Thickness estimation (0.50 mm³)
WM distance: 54s
CSF distance: 18s
PBT2x thickness: 56s
136s
Create initial surface 94s
Topology correction: 116s
Surface refinement: 87s
Reduction of surface collisions with optimization: 64s
Spherical mapping with areal smoothing 72s
Spherical registration 287s
rh:
Thickness estimation (0.50 mm³)
WM distance: 51s
CSF distance: 16s
PBT2x thickness: 54s
130s
Create initial surface 81s
Topology correction: 100s
Surface refinement: 91s
Reduction of surface collisions with optimization: 58s
Spherical mapping with areal smoothing 75s
Spherical registration 301s
Final surface processing results:
Average thickness (FS): 2.3629 ± 0.5880 mm
Surface intensity / position RMSE: 0.0774 / 0.0854
Euler number / defect number / defect size: 94.0 / 43.0 / 2.46%
Display thickness: C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\lh.thickness.Subject12
Display thickness: C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\rh.thickness.Subject12
Show surfaces in orthview: C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\Subject12.nii
Surface ROI estimation 13s
Surface and thickness estimation takes 1850s
ROI estimation in native space
ROI estimation of 'thalamic_nuclei' atlas 2s
ROI estimation of 'cobra' atlas 7s
ROI estimation of 'neuromorphometrics' atlas 24s
ROI estimation of 'lpba40' atlas 7s
ROI estimation of 'hammers' atlas 17s
ROI estimation of 'thalamus' atlas 2s
ROI estimation of 'suit' atlas 4s
ROI estimation of 'ibsr' atlas 6s
ROI estimation of 'aal3' atlas 10s
ROI estimation of 'mori' atlas 13s
ROI estimation of 'anatomy3' atlas 19s
ROI estimation of 'julichbrain' atlas 25s
ROI estimation of 'Schaefer2018_100Parcels_17Networks_order' atlas 12s
ROI estimation of 'Schaefer2018_200Parcels_17Networks_order' atlas 21s
ROI estimation of 'Schaefer2018_400Parcels_17Networks_order' atlas 46s
ROI estimation of 'Schaefer2018_600Parcels_17Networks_order' atlas 61s
Write results 64s
279s
Quality check 40s
C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\report\catreport_Subject12.pdf
------------------------------------------------------------------------
CAT preprocessing takes 55 minute(s) and 14 second(s).
Image Quality Rating (IQR): 70.35% (C-)
GM volume (GMV): 43.16% (689.58 / 1597.72 ml)
GM thickness (GMT): 2.36 ± 0.59 mm
Segmentations are saved in C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\mri
Reports are saved in C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\report
Labels are saved in C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\label
------------------------------------------------------------------------
14-Aug-2023 16:59:17 - Done 'CAT12: Segmentation'
14-Aug-2023 16:59:18 - Running 'Extract additional surface parameters'
1/1) Extract parameters for C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\lh.central.Subject12.gii
1s - Display C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\lh.gyrification.Subject12
6s - Display C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\lh.depth.Subject12
1s - Display C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\rh.gyrification.Subject12
6s - Display C:\Users\joerlema\.brainstorm\tmp\cat12_230814_160334\surf\rh.depth.Subject12
14-Aug-2023 16:59:33 - Done 'Extract additional surface parameters'
14-Aug-2023 16:59:33 - Done
BST> Processing dependencies: cat12 requires: spm12
BST> Plugin cat12 already loaded: C:\Users\joerlema\Desktop\ASO\Doctoraat\Faculty_of_Psychology\MATLAB\SPM\spm12\toolbox\cat12
BST> Saving protocol "Epilepsy_study"...
BST> Saving scouts in surface: Subject12/tess_lh.central.freesurfer.mat
BST> Saving scouts in surface: Subject12/tess_rh.central.freesurfer.mat
BST> Saving scouts in surface: Subject12/tess_lh.central.freesurfer_02.mat
BST> Saving scouts in surface: Subject12/tess_rh.central.freesurfer_02.mat
BST> Saving protocol "Epilepsy_study"...
BST> Saving protocol "Epilepsy_study"...
From the shared log, it seems there is a problem with the CAT12 that is being used.
This is not the CAT12 version that is installed by Brainstorm as plugin.
Can you try to remove SPM and CAT12 from the Matlab path and let Brainstorm to handle them as plugins?
I suspect there is something wrong with your installation of SPM/CAT. It looks like some of the installation folders are missing.
Make sure your SPM installation is also up-do-date.
If you are expecting to use CAT mainly from Brainstorm, it would be a safer to let Brainstorm install it:
Edit your MATLAB path: Remove all the references to SPM and CAT
Restart Matlab, make sure that typing "spm" or "cat12" in the Matlab command window return an error
Start Brainstorm, install SPM12 and CAT12 as…
Hello Raymundo,
I uninstalled and re-installed both SPM and CAT12 (however not as a plugin, because then I got the " error creating link"), and now it seemed to work. I don't really understand what I did differently compared to the previous subject, but fingers crossed I don't encounter this issue anymore.
Many thanks for your quick replies!
Kind regards,
Joyce
I'm glad it worked! It may be the case that either SPM or CAT12 were old versions that returned the data in an unexpected way.
joerlema:
I uninstalled and re-installed both SPM and CAT12 (however not as a plugin, because then I got the " error creating link"), and now it seemed to work. I don't really understand what I did differently compared to the previous subject, but fingers crossed I don't encounter this issue anymore.
It is strongly recommended to install SPM and CAT12 as plugins, in that way Brainstorm can have control on which scripts are on the Matlab path and which specific version of the plugins are run.
In order the issue you have with the plug in installation, please provide the following information:
The entire error text
Which OS you are using