Hello all,
I did something silly and changed the my channel file on my EEG CNT data in my common files from CNT 2D channels to a default Neuroscan 64 from Colin 27.
That has now locked me out of my data.
I have tried to reimport the specific .asc channel file i exported from my neuroscan system but it has failed to update in a manner that will allow my file to open.
ERROR
** Error: Line 375: bst_memory>LoadChannelFile (line 375)
** Number of channels in ChannelFile (64) and DataFile (65) do not match. Aborting…
**
** Call stack:
** >bst_memory.m>LoadChannelFile at 375
** >bst_memory.m>LoadDataFile at 584
** >bst_call.m at 26
** >macro_methodcall.m at 37
** >bst_memory.m at 71
** >view_timeseries.m at 78
** >tree_callbacks.m at 243
** >bst_call.m at 28
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 103
**
I tried exporting a .pos file and then updating that but i cant get the channel matrix to match.
Any thoughts?
Thank you
Josh
Hi Josh,
You can probably do something like this:
- Create a new subject (Default channel file: Yes, use one channel file per subject)
- Link you CNT file to this new subject (“Review raw file”)
- Copy-paste the new CNT channel file and replace the one that you modified - this one contains the correct list of channels
- Right-click on copied channel file > Add EEG positions
This is illustrated here:
http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Access_the_recordings
Cheers,
Francois
Hi Francois,
I tried this but it didnt allow me to use a file format that supported the .asc files or pasted files.
I also took the channel file from bs datatbase in the new subject from the original raw file i then substituted the default neuroscan channel file for the original file and reloaded the protocol.
Sadly it didnt work
below is the erro.
Any other thoughts?
Thank you
Josh
** Error: Line 835: Improper index matrix reference.
**
** Call stack:
** >panel_record.m>UpdatePanel at 835
** >panel_record.m>CurrentFigureChanged_Callback at 692
** >bst_call.m at 28
** >macro_methodcall.m at 39
** >panel_record.m at 28
** >bst_figures.m>SetCurrentFigure at 975
** >bst_call.m at 28
** >macro_methodcall.m at 39
** >bst_figures.m at 59
** >view_timeseries.m at 191
** >tree_callbacks.m at 243
** >bst_call.m at 28
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 103
**
BST> Error: no data could be found for this figure...
I tried this but it didnt allow me to use a file format that supported the .asc files or pasted files.
Can you please post here your .asc file? or copy-paste it in a post (between the tags "[ CODE ]file contents[ /CODE ]")
I also took the channel file from bs datatbase in the new subject from the original raw file i then substituted the default neuroscan channel file for the original file and reloaded the protocol.
I'm not sure I understand what you describe here. Can you be more precise?
Hi Francois
.asc is an invalid file to post si i have copied it here.
;SCAN 4.0 ASCII Channel Description File
;
;Line format:
;#<channel number> <label>
;or:
;<display page> <channel number> <pos x0> <pos y0> <pos x1> <pos y1>
;
;A semicolon at the first column indicates a comment.
;
#1 FP1
#2 FPZ
#3 FP2
#4 AF3
#5 AF4
#6 F7
#7 F5
#8 F3
#9 F1
#10 FZ
#11 F2
#12 F4
#13 F6
#14 F8
#15 FT7
#16 FC5
#17 FC3
#18 FC1
#19 FCZ
#20 FC2
#21 FC4
#22 FC6
#23 FT8
#24 T7
#25 C5
#26 C3
#27 C1
#28 CZ
#29 C2
#30 C4
#31 C6
#32 T8
#33 TP7
#34 CP5
#35 CP3
#36 CP1
#37 CPZ
#38 CP2
#39 CP4
#40 CP6
#41 TP8
#42 P7
#43 P5
#44 P3
#45 P1
#46 PZ
#47 P2
#48 P4
#49 P6
#50 P8
#51 PO7
#52 PO5
#53 PO3
#54 POZ
#55 PO4
#56 PO6
#57 PO8
#58 O1
#59 OZ
#60 O2
#61 M1
#62 M2
#63 SSTpulse
#64 GFP
#65 REF
0 1 0.385 0.099 0.423 0.145
0 2 0.483 0.093 0.520 0.139
0 3 0.580 0.099 0.617 0.145
0 4 0.385 0.200 0.423 0.246
0 5 0.577 0.188 0.615 0.233
0 6 0.229 0.230 0.266 0.276
0 7 0.291 0.273 0.328 0.319
0 8 0.356 0.288 0.392 0.334
0 9 0.421 0.297 0.457 0.343
0 10 0.483 0.285 0.520 0.331
0 11 0.548 0.291 0.585 0.337
0 12 0.612 0.288 0.650 0.334
0 13 0.675 0.270 0.712 0.316
0 14 0.737 0.230 0.774 0.276
0 15 0.188 0.343 0.225 0.389
0 16 0.266 0.374 0.303 0.420
0 17 0.342 0.386 0.380 0.432
0 18 0.415 0.386 0.452 0.432
0 19 0.483 0.380 0.520 0.426
0 20 0.556 0.383 0.593 0.429
0 21 0.626 0.380 0.663 0.426
0 22 0.702 0.371 0.739 0.416
0 23 0.785 0.346 0.823 0.392
0 24 0.172 0.484 0.209 0.529
0 25 0.264 0.481 0.301 0.526
0 26 0.339 0.474 0.376 0.520
0 27 0.415 0.471 0.452 0.517
0 28 0.485 0.468 0.523 0.514
0 29 0.556 0.468 0.593 0.514
0 30 0.629 0.471 0.666 0.517
0 31 0.704 0.471 0.742 0.517
0 32 0.791 0.474 0.828 0.520
0 33 0.191 0.615 0.228 0.661
0 34 0.275 0.581 0.311 0.627
0 35 0.348 0.563 0.384 0.609
0 36 0.418 0.554 0.455 0.600
0 37 0.485 0.551 0.523 0.597
0 38 0.556 0.551 0.593 0.597
0 39 0.623 0.554 0.660 0.600
0 40 0.694 0.569 0.731 0.615
0 41 0.777 0.600 0.815 0.645
0 42 0.248 0.722 0.284 0.767
0 43 0.307 0.676 0.344 0.722
0 44 0.366 0.645 0.404 0.691
0 45 0.423 0.633 0.460 0.679
0 46 0.485 0.636 0.522 0.682
0 47 0.548 0.633 0.585 0.679
0 48 0.607 0.642 0.645 0.688
0 49 0.666 0.670 0.704 0.716
0 50 0.729 0.703 0.766 0.749
0 51 0.312 0.795 0.350 0.841
0 52 0.350 0.758 0.387 0.804
0 53 0.393 0.731 0.431 0.777
0 54 0.488 0.725 0.525 0.771
0 55 0.583 0.725 0.620 0.771
0 56 0.626 0.746 0.663 0.792
0 57 0.661 0.786 0.698 0.832
0 58 0.402 0.832 0.439 0.877
0 59 0.488 0.828 0.525 0.874
0 60 0.577 0.825 0.615 0.871
0 61 0.064 0.688 0.101 0.734
0 62 0.907 0.667 0.944 0.713
0 63 0.123 0.023 0.160 0.069
0 64 0.703 0.832 0.803 0.932
0 65 0.746 0.832 0.846 0.932
1 1 0.028 0.032 0.139 0.161
1 2 0.167 0.032 0.278 0.161
1 3 0.306 0.032 0.417 0.161
1 4 0.444 0.032 0.556 0.161
1 5 0.583 0.032 0.694 0.161
1 6 0.722 0.032 0.833 0.161
1 7 0.861 0.032 0.972 0.161
1 8 0.028 0.194 0.139 0.323
1 9 0.167 0.194 0.278 0.323
1 10 0.306 0.194 0.417 0.323
1 11 0.444 0.194 0.556 0.323
1 12 0.583 0.194 0.694 0.323
1 13 0.722 0.194 0.833 0.323
1 14 0.861 0.194 0.972 0.323
1 15 0.028 0.355 0.139 0.484
1 16 0.167 0.355 0.278 0.484
1 17 0.306 0.355 0.417 0.484
1 18 0.444 0.355 0.556 0.484
1 19 0.583 0.355 0.694 0.484
1 20 0.722 0.355 0.833 0.484
1 21 0.861 0.355 0.972 0.484
1 22 0.028 0.516 0.139 0.645
1 23 0.167 0.516 0.278 0.645
1 24 0.306 0.516 0.417 0.645
1 25 0.444 0.516 0.556 0.645
1 26 0.583 0.516 0.694 0.645
1 27 0.722 0.516 0.833 0.645
1 28 0.861 0.516 0.972 0.645
1 29 0.028 0.677 0.139 0.806
1 30 0.167 0.677 0.278 0.806
1 31 0.306 0.677 0.417 0.806
1 32 0.444 0.677 0.556 0.806
1 33 0.583 0.677 0.694 0.806
1 34 0.722 0.677 0.833 0.806
1 35 0.861 0.677 0.972 0.806
1 36 0.028 0.839 0.139 0.968
1 37 0.167 0.839 0.278 0.968
1 38 0.306 0.839 0.417 0.968
1 39 0.444 0.839 0.556 0.968
1 40 0.583 0.839 0.694 0.968
Regarding the channel file.
I found the Neuroscan quickcaps channel .mat file in the brainstorm database folder which I then removed and added the new channel .mat file from the new subject file i loaded in with the linked raw file.
I hope this is clearer.
Cheers
Josh
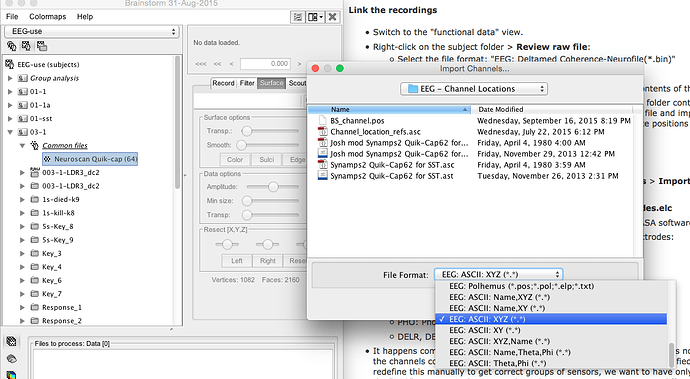
Hi Josh,
You can import this .asc file with the file format “EEG: Neuroscan”, but it doesn’t contain any 3D positions, just a flat 2D display map.
To add the default 3D positions from Brainstorm to your own files, you should be able to do the following:
- Link you CNT file in the database (“Review raw file”)
- Right-click on the channel file created automatically > Add EEG positions > Default: Colin27 > Neuroscan Quik-cap 64
Francois
Thank you Francois for your help.
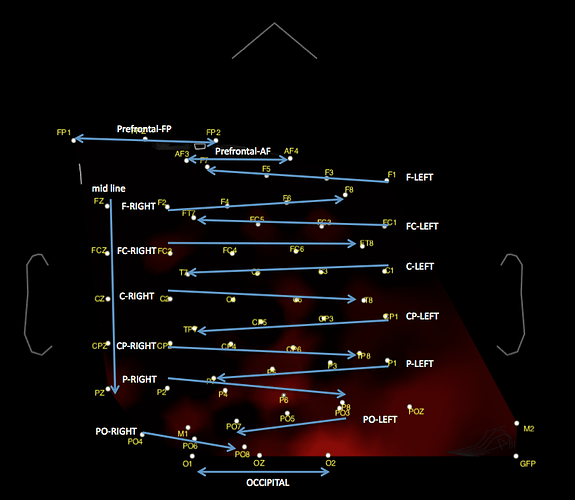
I have another problem with the eeg montage that you may be able to help with. I added a global rereference channel in the neuroscan edit suite and that has appeared to thrown off the montage completely (See attached image)
I have included an image of how the montage got skewed. I am running a 65 channel montage from a neuroscan 64 channel quickcap system. However when i change the channel file to the 64 channel neuroscan montage it will not let me open up the file. Do you have any advice?
Many thanks
Josh
I’m not sure I understand what you explain.
If you follow exactly the procedure described in my previous message, I don’t think you can have the problems you describe:
- Link you CNT file in the database (“Review raw file”)
- Right-click on the channel file created automatically > Add EEG positions > Default: Colin27 > Neuroscan Quik-cap 64
If you really don’t understand where the error is coming from:
- Send me the files you are trying to import
- Describe step by step the operations you are doing in Brainstorm, with screen captures
- Describe what is not working and is it different from what you expect (with screen captures too).
Francois