Hi
I’ve tried to import channel location to brainstorm but unfortunately, I have this error:
No channel matching the loaded cap.
I have this problem when I use my EEG file with .mat format while using the .eeg (BrainVision Recorder) format I don’t have this problem.
I’ve tried with several channel location format such as .txt, .sfp, .mat but all of them don’t work.
I need to use my .mat file would you please let me know how can set my channel location?
I uploaded my EEG and location files in this address.
https://ufile.io/zt4gp
Best Regards,
Hamed
Hi,
Your uploaded SFP file can be imported into Brainstorm by right clicking on your subject -> Import channel file -> EEG: EGI (*.sfp). As for your MAT file, it can be imported by right clicking on your subject -> Import MEG/EEG -> EEG: Matlab matrix (*.mat).
It seems your channel file contains 65 channels whereas your Matlab file has data for 64 of them. Right click on your EGI channels once imported and select Edit channel file and change the type to the unneeded one to (Delete) to avoid problems.
Cheers,
Martin
Hi Dear Martin
Thanks a lot for your reply.
yes, that's works. but I have a new problem with this channel locations.
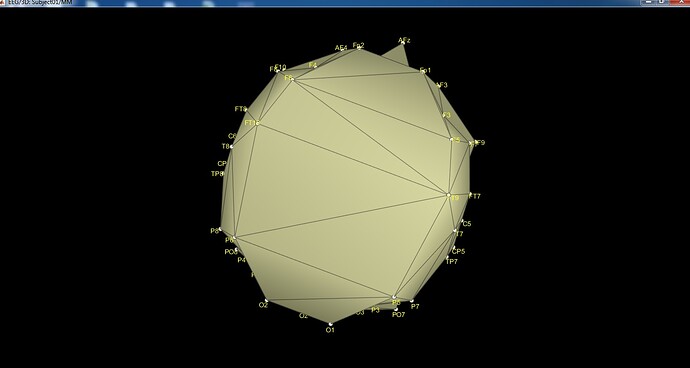
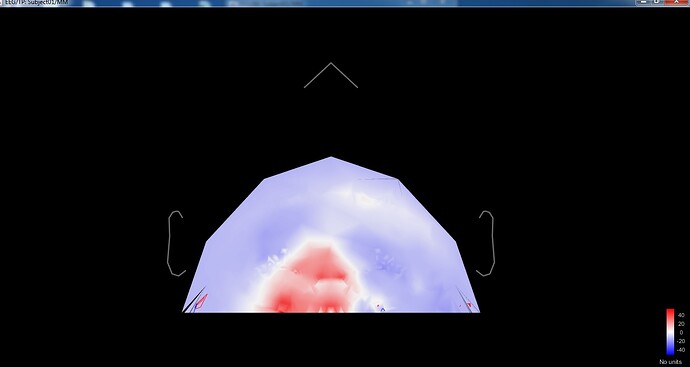
When I want to see the channel location in 2D and 3D I saw an image that it's not normal and related to my channel location.
and I have an error about the same position of channels too but the positions are ok. I've tested in EEGLAB and it's ok there.
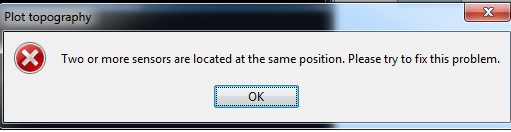
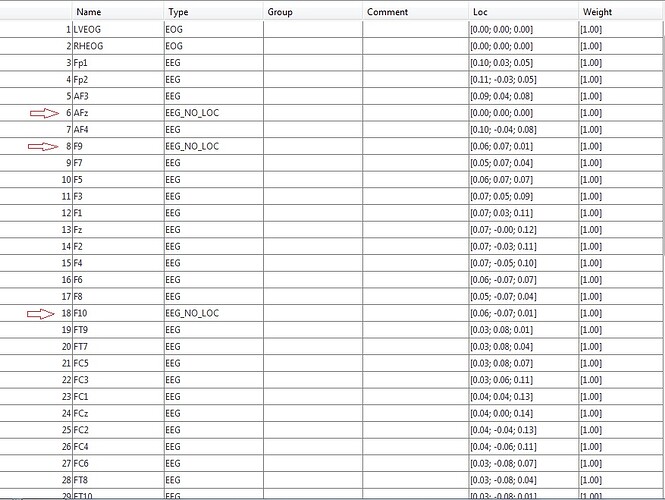
@Francois any ideas? It seems point FC3 = FC5 and point FC4 = FC6, yet in the SFP files they have different coordinates.
Hi Francois
Thanks a lot for your reply.
First of all, I should say our EEG cap is ActiCap 64 ch
1- I don't know why they used commas. I downloaded from EasyCap website:
http://www.easycap.de/e/downloads/M1_XYZ.txt
2- yes there is a standard position and I removed some of the channels that I don't need them.
3- When I use the BrainStorm channel location there is ActiCap 65 no Acticap64.
and when I use ActiCap65 it can't recognize my 3 channels (AFz, F9, F10).
and another question is when I exported my analyzed file to Matlab how can I access directly to my file.
I've searched in all cells but I couldn't find them in Row and Column format (my file :64*34159).

Best Regards,
Hamed
Hi Hamed,
The cap you describe here is referenced in Brainstorm as “EasyCap M1”, not as “ActiCap 64”. Select this entry and it should work correctly.
The file you are looking at is the link to the continuous file:
http://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#On_the_hard_drive
The data is imported and copied to your database only when you epoch it:
http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#On_the_hard_drive
If you are not familiar with these notions, I recommend you start by reading all the introduction tutorials in the “Get started” section of the website.
Cheers,
Francois
Hi Francois
Thanks a lot, it works.
For exporting .mat file I’ll read the tutorial
Thanks again
Cheers,
Hamed