Hello Francois, I was hoping you could check over my source script to see if I followed the directions right. Here’s a little info about the EEG data. The EEG data was all pre-processed (cleaned) in eeglab and is then imported into brainstorm. It is a continuous file of 3 1 minute recordings of pre stimulation baseline, during stimulation period, and baseline post stimulation period. No individual MRIs were used. We are using brainstorms default head model.
Here is my source script
- Create new protocol
a. Yes use protocols default anatomy
b. No, use one channel file per acquisition run - Create subject (under anatomy tab)
a. Yes, use protocols default anatomy
b. No, use one channel file per acquisition run - Import MEG/EEG (click on subject and import MEG/EEG)
a. EEG scaling factor set to 10 mm
b. Import EEG file since this is a continuous data file I unselect “use events”, but click on “create a separate folder for events”
c. (Sensors/anatomy registration) refine registration now? YES
It still looks odd
d. To fix this I click on EEG lab channels--> MRI registration --> edit --> head pops up and I click on “project electrodes to surface” and it looks perfect.
-
Create head model --> Click back on subject and click “create head model” I check if the fiducials are correct and they always are by default I think? I’ve never had to move them. Question: If you are using the default head model, would you ever need to change the fiducials?
a. OpenMEEG BEM pops up and I click --> cortex surface and forward modeling methods “openMEEGBEM”
b. For BEM layers and conductives I leave everything at default -
Create Sources
a. I click on the data set folder and click --> noise covariance --> no -noise modeling, since all of my EEG imported is meaningful data
b. I click back on the data set folder and click --> compute sources 2018 --> since I am using continuous data (non-focal data) I use min norm imaging --> for measure I use sloreta . Question: Is it ok to use min norm imaging and sLORETA with no noise data covariance? -
Extracting source data
a. To extract my source data I click on my raw file --> move to the files to process box --> click on the source brain on the left and then run
b. Pipeline editor process section pops up --> extract
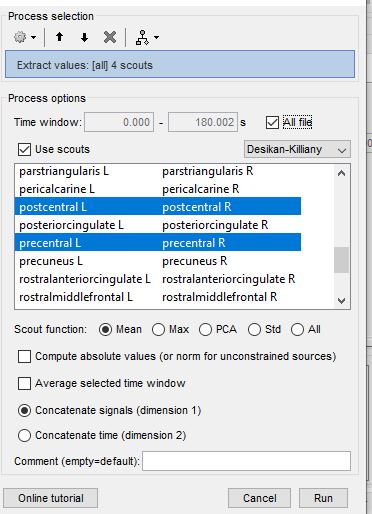
c. Lastly here a pic of what I select. Question: If im using 1 continuous file, I don’t understand the difference between concatenate signals (dimension 1) vs concatenate time (dimension 2). Both options give me the same values. Im assuming this has something to do with ERPS
d. After I extract my values I do further analysis in matlab and treat this source date the same exact way as EEG data when applying these matlab scripts for different analysis (wavelet transformation, power analysis.. partial directed coherence etc….)
Im hoping this is all correct? Thanks for your time! Sorry this is so long.