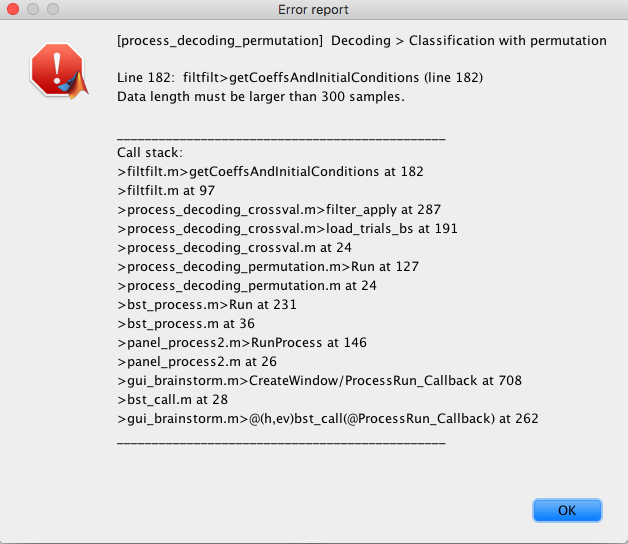
Hello,
Apologies if this has been posted before, but I'm having some trouble using the decoding / classification process. I followed the tutorial from this page (http://neuroimage.usc.edu/brainstorm/Tutorials/Decoding) successfully with no issues, but I've run into the error when trying to use this on my own data-set.
A few other possibly relevant notes:
-
The data seems fine, but it did undergo several conversion processes from another file format to get it Brainstorm ready (it's now in the ".set" file format, which is also used in EEGLAB); I don't currently have names for the channels, nor is the data calibrated/normalized, not sure if that matters.
-
I haven't yet run any sort of artifact detection/cleaning yet - this was purely a preliminary move to see what the classifier output.
I'm getting the error when I run both cross-validation or permutation . I have 36 samples from each condition. Any idea what this is from?
Let me know if you need more information!
Thanks,
Sean Trott