Dear Brainstorm,
I'm currently doing group-level statistics on MEG volume-based source data. My question is whether it is appropriate to use cluster-based permutation (process_ft_sourcestatistics) for the statistical analysis of volume-based source data.
Background: In my study, I have found a significant difference between two conditions in the high beta band (0-168 ms, 22-30 Hz) at the sensor level using the cluster-based permutation. Next, I want to identify the specific brain regions that account for this difference between the two conditions at the source level. So, I processed "volume source estimation" with the following steps:
Generate source grid
Compute a head model using this template grid
Compute source [2018]: dSPM, Unconstrained, MEG sensors
Hilbert transform: high beta, '22, 30', 'mean'
Apply a baseline norm: db correction
Project on default anatomy
Resample data to 250 Hz
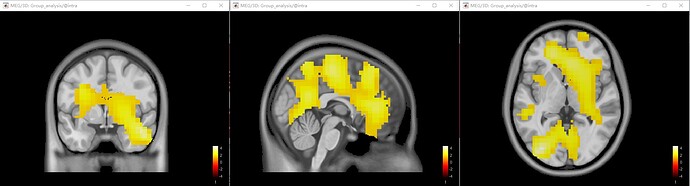
Cluster-based permutation (process_ft_sourcestatistics) : 0-168 ms averaged time.
The result was a large and significant cluster (P = 0.032) including some subcortical regions and so on.
I think this cluster is very large and includes many brain areas, so I'm not sure if I made a mistake in my data analysis and whether it is acceptable to use cluster-based permutation to analyze volume-based source data. After searching related literature, I have yet to find any references for this method. Could anyone provide me with references or guidance on this topic?
Best,
Shuting