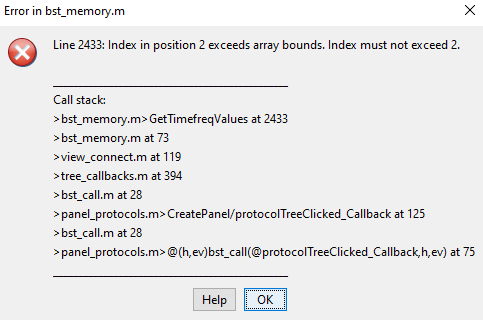
Hello,
I am trying to write my script to automate the process of importing the data into brainstorm, setting the channel file, and lastly find the PLV values. I remember reading somewhere that importing the data should be done in one process and have another process for PLV. Is there a way I can implement this strategy of of separate processes into my code? I mean could anyone explain why I keep getting errors for the code shown below even though it works fine for other functional connectivity metrics such as coherence and correlation.
The portion I am talking about in my current code that is giving me an error:
%% Basic processes
% Process: Import MEG/EEG: Existing epochs
sFiles = bst_process('CallProcess', 'process_import_data_epoch', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'datafile', {data_file_to_use,'FIF'}, ...
'iepochs', [], ...
'eventtypes', '', ...
'createcond', 0, ...
'channelalign', 1, ...
'usectfcomp', 1, ...
'usessp', 1, ...
'freq', [], ...
'baseline', [], ...
'blsensortypes', 'EEG');
% Process: Set channel file
sFiles = bst_process('CallProcess', 'process_import_channel', sFiles, [], ...
'channelfile', {RawFiles{current_length}, 'ASCII_NXYZ'}, ...
'usedefault', 1, ... %
'channelalign', 1, ...
'fixunits', 1, ...
'vox2ras', 1);
% Process: Phase locking value
sFiles = bst_process('CallProcess', 'process_plv1n', sFiles, [], ...
'timewindow', [0, 15], ...
'dest_sensors', 'EEG', ...
'includebad', 1, ...
'plvmethod', 'plv', ... % 'plv' %'ciplv' %'wpli'
'plvmeasure', 2, ... % Magnitude
'tfmeasure', 'hilbert', ... % Hilbert transform
'tfedit', struct(...
'Comment', 'Complex', ...
'TimeBands', [], ...
'Freqs', {{ 'beta', '13, 29', 'mean'}}, ...
'ClusterFuncTime', 'none', ...
'Measure', 'none', ...
'Output', 'all', ...
'SaveKernel', 0), ...
'timeres', 'windowed', ... % Full (requires epochs)
'avgwinlength', 1, ...
'avgwinoverlap', 50, ...
'outputmode', 'input'); % separately for each file
num_runs=length(sFiles);
for run=1:num_runs
current_folder_name={sFiles(run).FileName};
current_matrix= in_bst_matrix(current_folder_name{1});
TF_time_avg=mean(current_matrix.TF,3);
current_matrix.TF=TF_time_avg;
R = bst_memory('GetConnectMatrix', current_matrix); % get full matrix
feature_extraction_plv(R,case_index,num_runs,run);
end
end