Hello there,
I'm new to Brainstorm, and can't seem to find a way to calculate EOG channels - in BrainVision Analyzer, which I used before, this was easily achieved using the Linear Derivation transformation, and choosing the channels to be re-referenced to each other (e.g. (fp1 + fp2)/2 = VEOG).
Is there any way to achieve this is Brainstorm?
Thanks ahead for any help,
Nick
Hi Nick:
It’s actually possible. We call this forming ‘clusters’ of sensors. When viewing your data time series, highlight the sensors you want to group as (fp1 + fp2)/2 = VEOG, or using the ‘Clusters’ panel of the main interface, type in the name of the sensors you want to group, ie fp1 and fp2 and make sure the ‘Mean’ function is selected (that’s the default operation on clusters of sensors, but others are possible e.g., PCA, etc.). Name this cluster VEOG through the interface and save it, all using the GUI, for future use.
There is a bit of documentation about that: http://neuroimage.usc.edu/brainstorm/Tutorials/TutClusters?highlight=(cluster), which is for the online tutorial.
Let us know if this is helping or if you’d like to see more features added.
Dear Sylvain,
Thanks for the quick response. I tried using the “clusters” option, but this only seems to give a new display, not a new channel. What I really want to do is create two new bipolar eye channels (HEOG, VEOG), by combining existing regular ones. So, for example, the new VEOG channel would be obtained by taking channel Fp1 minus Fp2 (thus creating a new bipolar channel, wherein all common activity of the two old channels is subtracted away, leaving only the difference). Similarly, HEOG would be a linear derivation from F9 and F10.
In the end, these would be used to detect and remove eye blinks (which I can’t do now with just the sensor “cluster” - it is not even visible in the Artifacts>Detect Eye Blinks menu).
I hope this can be achieved in Brainstorm.
Let me know,
Nick
Dear Nick,
No, I’m sorry there is no way yet to either create a new channel from a linear combination of existing channels, or to perform the detection from a group of sensors (“cluster”).
I’m adding this to the list of features to add to Brainstorm, but it will take a long time before we get there.
Maybe you can find a way to perform this operation with you EEG acquisition software, to calculate the new channels before you import you recordings in Brainstorm.
You also have ways to do this by manually editing the files in the Brainstorm database, but it is not such an easy task, and requires some Matlab coding.
Let me know if you want more information about this.
Francois
Dear Francois,
Thank you so much for your speedy response. I’m sorry to hear Brainstorm can’t create new channels by derivation from existing ones.
I’m not sure I will be able to perform the operation prior to the import stage, so I’d be willing to try and make the changes to the database files you mentioned. Truth be told, I have no experience coding in Matlab, but if you would give me detailed instructions on how to edit the files, I would do it…
Thanks again Francois! I look forward to your reply,
N
Ok, here is the procedure.
You need to update all the data files (=recordings) and channel files.
You can either load/save them using directly matlab commands, if you want to do with a script, or using the interface.
To modify a file using the GUI:
- right-click on the file > File > Export to Matlab > “fileMat”
- change one or more fields of the structure “fileMat” from your Matlab command window
- right-click on the same file > File > Import from Matlab > “fileMat”
In the channel file:
- Add a new channel in the “Channel” field:
> iChannel = length(fileMat.Channel) + 1;
> fileMat.Channel(iChannel).Comment = ‘new_channel_name’;
> fileMat.Channel(iChannel).Type = ‘new_channel_type’;
> fileMat.Channel(iChannel).Loc = [0;0;0];
> fileMat.Channel(iChannel).Weight = 1;
- If you have some SSP projectors defined, you need to add an empty row at the bottom of each fileMat.Projector(i).Components entry
In the data file:
- Add a row at the bottom of the F matrix, as a combination of channels #i1 and #i2
> fileMat.F(iChannel, = fileMat.F(i1) + fileMat.F(i2);
= fileMat.F(i1) + fileMat.F(i2);
- Add an entry in the ChannelFlag field (the bad channels are tagged with a -1 in this vector, good channels are set to 1):
> fileMat.ChannelFlag(iChannel) = 1;
Notes:
- you won’t be able to use those file to run the automatic blink detection, as it works only on continuous native files for now.
- this works only on imported data blocks, on the “Link to raw files”.
Good luck
Francois
[QUOTE=Francois;4480]Ok, here is the procedure.
You need to update all the data files (=recordings) and channel files.
You can either load/save them using directly matlab commands, if you want to do with a script, or using the interface.
To modify a file using the GUI:
- right-click on the file > File > Export to Matlab > “fileMat”
- change one or more fields of the structure “fileMat” from your Matlab command window
- right-click on the same file > File > Import from Matlab > “fileMat”
In the channel file:
- Add a new channel in the “Channel” field:
> iChannel = length(fileMat.Channel) + 1;
> fileMat.Channel(iChannel).Comment = ‘new_channel_name’;
> fileMat.Channel(iChannel).Type = ‘new_channel_type’;
> fileMat.Channel(iChannel).Loc = [0;0;0];
> fileMat.Channel(iChannel).Weight = 1;
- If you have some SSP projectors defined, you need to add an empty row at the bottom of each fileMat.Projector(i).Components entry
In the data file:
- Add a row at the bottom of the F matrix, as a combination of channels #i1 and #i2
> fileMat.F(iChannel, = fileMat.F(i1) + fileMat.F(i2);
= fileMat.F(i1) + fileMat.F(i2);
- Add an entry in the ChannelFlag field (the bad channels are tagged with a -1 in this vector, good channels are set to 1):
> fileMat.ChannelFlag(iChannel) = 1;
Notes:
- you won’t be able to use those file to run the automatic blink detection, as it works only on continuous native files for now.
- this works only on imported data blocks, on the “Link to raw files”.
Good luck
Francois[/QUOTE]
Oh, so this procedure wouldn’t help me with automatic blink detection. I better not mess with the source files then, since blink algorithms were the main reason I wanted to do it…
One workaround would be if I could do this in EEGlab and then import thee eeglab file into BST?
Hello:
You may want to try the automatic processes Brainstorm has for blink detection and correction by selecting directly a frontal channel that is strongly contaminated by blinks in your data. It might be good enough. See documentation at http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?highlight=(blink)#Automatic_detection:_vEOG
An alternative is indeed to proceed as you wish in EEGLab and then import the clean data and/or the blink events back into Brainstorm (which can read EEGlab data format)
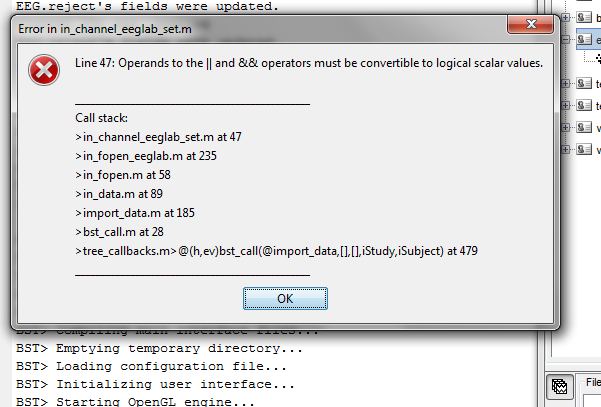
Hi Sylvain,
When I try to import the EEGlab .set file, I get the following error screen:
Hello
I think I fixed this bug.
Can you update Brainstorm (menu Help > Update Brainstorm) and try again?
Francois
Updating Brainstorm fixed the bug indeed. Thanks, both!
Hi!
I hope you guys are ok and your families are safe!
I just want to ask : in the current BS version is there any way to create a new channel with some electrode combinations?
Thanks in advance!
Create a custom montage, then use the process "Standardize > Apply montage".
https://neuroimage.usc.edu/brainstorm/Tutorials/MontageEditor#Custom_montage
https://neuroimage.usc.edu/brainstorm/Tutorials/MontageEditor#Apply_a_montage_to_the_recordings
Please try to post your questions at the end of threads that are strictly related with your question, otherwise create a new topic. Thanks.
 = fileMat.F(i1) + fileMat.F(i2);
= fileMat.F(i1) + fileMat.F(i2);