Hi there,
We noticed that when calculating the AEC on EEG data the diagonal of the matrix has values >0.2, why is that?
When we visualize the matrix in Brainstorm it seems like the diagonal is zero (the plotting function sets it to zero) but when we export the data and use them for our analysis the diagonal is far from zero. Can you explain why that is the case? How should we treat those values?
We export the matrix using the following:
bst_memory('GetConnectMatrix', conn_file_extracted_from_Brainstorm)
Hi @eletamilia,
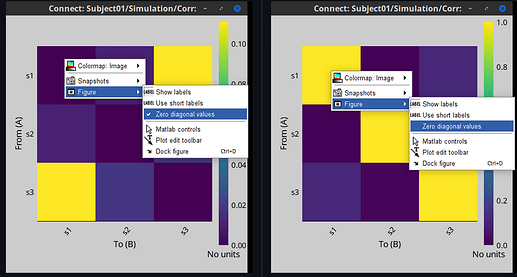
The diagonal elements are forced to zero so only for display, so the off-diagonal values can be visualized properly instead of being scaled by the large values obtained in the connectivity matrix diagonal.
The exported matrix will have the same [Na x Nb x Ntime x Nfreq]
More information about this here: https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#On_the_hard_drive
Lastly, today we added the option to make optional the zeroing of the diagonal when visualizing connectivity matrices. To get this option, just update Brainstorm.
Best,
Raymundo
Thanks for your reply.
My concern or question though is more: Why are the diagonal values different from zero? That is what is unclear to me. What are those values?
Those diagonal values correspond to the connectivity measure of a signal with itself.
Which process you using for AEC?
Note that few months ago the Amplitude Envelope Correlation processes has been deprecated, use instead the processes Envelope Correlation [2022]
Best,
Raymundo