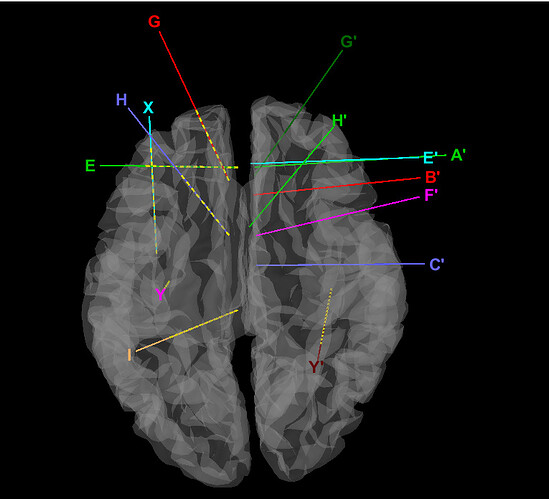
Hi, I'm locating SEEG,some of the contacts do not show, and the positions of them are also [0, 0, 0] in the channel file. Some shafts of this subject are OK, some are not. Anyone knows why?
Please describe better your starting point (what you had in the beginning) and what you did in Brainstorm (all the clicks step by step).
Hi, I'm locating the SEEG with pre-T1 MRI and post CT. After co-registering them, I imported SEEG data, checked the group of each contact, then located them. Firstly I clicked the top of the shaft, then the skull entry(of course the contact spacing is correctly set).
Can you please share your channel file?
(upload the channel_...mat file somewhere and post the download link here)
Here it is. It's in NAME_XYZ_MNI format.
And you could see some of the shafts show no contacts. This is pretty odd, for most of the contacts on the other side of the brain are normal.
I meant the actual channel_...mat from the Brainstorm database.
To find it: Right-click on the channel file > File > Show in file explorer.
Sorry, my bad
Have you imported the eletrodes positions for a text file such as the sample.txt file you posted earlier?
If this is the case: there are many contacts missing in this file...
To fix your current channel file:
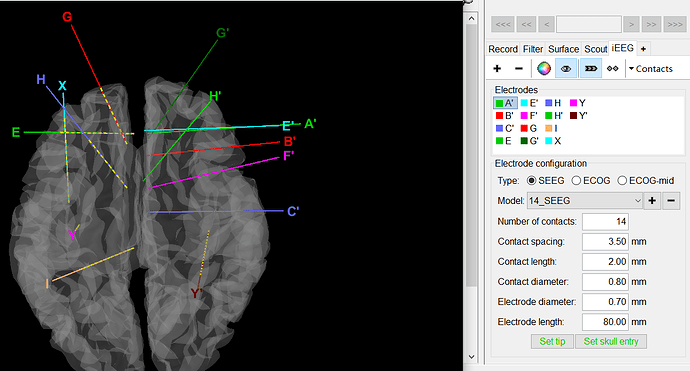
- Right-click on the channel file > Edit channel file > Select all the channels that have the type "SEEG_NO_LOC" (the contacts missing in the position file you tried to import), right-click > Set channel type > Enter "SEEG". Close the window to save the modifications.
- Double-click on the channel file to open it. In the tab iEEG: select all the electrodes with missing contacts, and click on the menu Contacts > Use default positions.
I did firstly import a channel file.
Problem solved! Thank you!