Thank you for this very helpful clarification.
How can we automatically create the NAS/LPA/RPA points? Is it done using the MNI normalization?
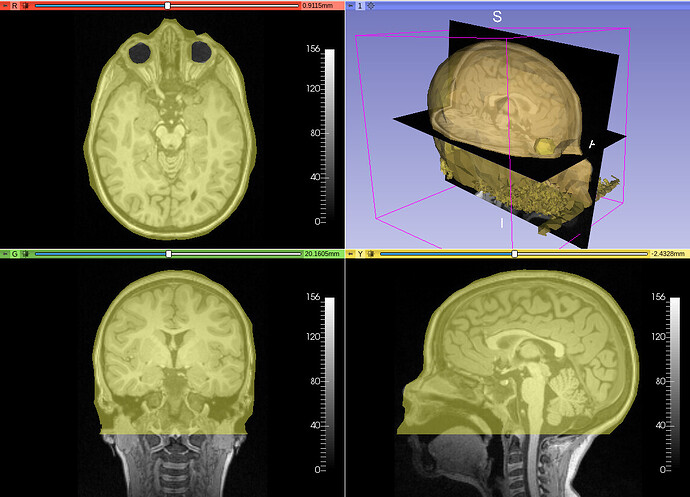
I was able to succesfully create a FEM mesh that corresponds to the images in 3D Slicer using the following steps:
-
Create a new subject using "individual anatomy".
-
Import the T1 MRI in niftii format, applying the image transform.
-
Perform MNI normalization of the T1 MRI to Create SCS coordinate system using the maff8 method.
-
Import the T2 MRI, applying the image transform, registering it to the T1 MRI using the SPM method and reslicing this volume. Without registration and resampling the brain2mesh step fails when using both the T1 and T2 MRIs.
-
Select both the T1 and T2 MRIs and generate the FEM mesh using the brain2mesh method.
-
Export the mesh and T1 MRI to Matlab variables called 'mesh' and 'mri'.
-
Convert the coordinates from SCS to world coordinates using this code:
[mesh.Vertices, Transf] = cs_convert(mri, 'scs', 'world', mesh.Vertices(:,1:3)); -
Scale the coordinates of the mesh using this code:
mesh.Vertices = 1000 * mesh.Vertices -
Save the mesh to Gmsh format using this code:
out_fem_msh(mesh, 'mesh.msh') -
Open "mesh.msh" in Gmsh and export to "mesh.vtk".
-
Open "mesh.vtk" in Slicer using RAS coordinate system.
The mesh overalyed on the T1 MRI looks like this: