Hello,
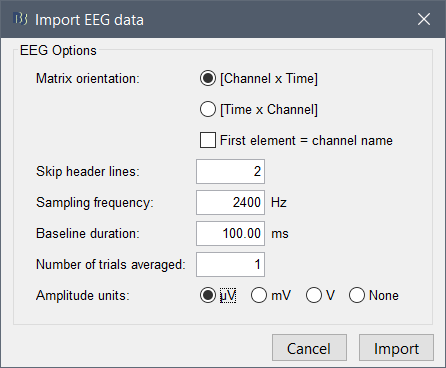
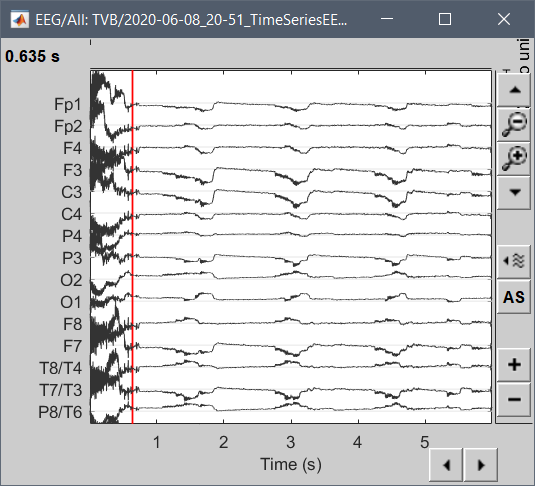
I am trying to import simulated EEG data produced by The Virtual Brain (TVB) into brainstorm for analysis. The simulated data follows the standard 10-20 montage with 63 surface electrodes. Any data exported from TVB comes out in a .h5 file. I can read these in Python using h5py.File('filename.h5', 'r') and have determined that the simulated EEG data consists of 2 datasets, ['data', 'time']. Viewing each individually reveals that data is of shape (6000, 2, 63, 1), while time is of shape (6000,). For specifics, I have included the code I ran and the subsequent output below.
The code:
import h5py
file = h5py.File('filename.h5','r')
print('file name')
print(file.name)
print('file keys')
print(file.keys())
print('structure of data')
print(file['/data'])
print('structure of time')
print(file['/time'])
The output:
file name
/
file keys
<KeysViewHDF5 ['data', 'time']>
structure of data
<HDF5 dataset "data": shape (6000, 2, 63, 1), type "<f8">
structure of time
<HDF5 dataset "time": shape (6000,), type "<f8">
All the data is there but I am unsure how to put it into a format that I can import into Brainstorm. The data is structured like a numpy array. I can get the first row of data with file['/data'][0, :, :, :]
Additionally, if anyone can help me get the head model data from TVB into Brainstorm, that would be extremely helpful. I am not the one who created the simulation in TVB and have not found the MRI data as of yet. However, both Brainstorm and TVB can import data from FreeSurfer and this appears to be the preferred method to prepare head model data for both. From TVB I can export .h5 files containing the cortical surface, face surface, brain skull interface, skin air interface, and skull skin interface data. The .h5 file containing the cortical surface data contains the following datasets ['triangle_normals', 'triangles', 'vertex_normals', 'vertices'] and was prepared using the following scripts (available on GitHub: https://github.com/timpx/scripts).
If anyone has recommendations or advice on either point it would be greatly appreciated. Stay safe and healthy.