I ran loadh5() on the .h5 file from TVB and it returned the following:
data: [1×63×2×6000 double]
time: [6000×1 double]
This is OK, but not sufficient.
You need to find a file that tells you at least the name of these 63 electrodes.
Do these files have standardized names?
Could you point at some online documentation describing this file format / the way to access these files?
It would be useful to invest time in writing a proper TVB .h5 reader in Brainstorm only if any TVB user could benefit from it in an easy way.
If this is the case, please send me a short example file (data + channel names), I'll see how I can organize this.
Otherwise, you can import these files in Brainstorm with the following procedure:
- Permute the dimensions to have [Nchannels, Ntime, Nepochs]:
test.data = permute(test.data, [2,4,3,1]) - Save the structure as a .mat file:
save('test_file.mat', '-struct', 'test') - Right-click on your subject in the database > Import MEG/EEG > Select file format "EEG: Matlab matrix (.mat)", select the file test_file.mat
- Configure the import so that it matches what you have in the file (the field "time" will be ignored and reconstructed from the information you set in the import options):
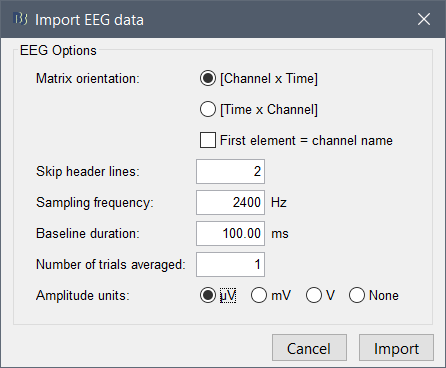
- Right-click on the channel file > Edit channel file : Edit the types and names of the channels