Hello,
I'm trying to import co-ordinates for SEEG electrodes. These have already been made in Slicer (which perhaps uses a difference space/matrix/coordinate system to Brainstorm/SPM/Matlab.
Any idea how to resolve this problem? How do i decide what transformation matrix to use?
Aswin
Hello,
I need to add something that is still missing: importing electrode positions in "world" coordinates:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems#World_coordinates
I will try to work on this within the next few days.
Don't hesitate to send me a reminder if this is not done next week.
Francois
Hello,
I finally added this option to add explicitly coordinates in "world" coordinates:
https://github.com/brainstorm-tools/brainstorm3/commit/59c2b65a65f3a0a16c2c73b77806c900b8d46743
Link your SEEG recordings to your Brainstorm database, right-click on the channel file > Add EEG positions > Load from file > Select one of the EEG: ASCII formats ending with "_World" (N=Name), depending on the format of the file you have created from slicer.
This would work only if the positions are exported based on the transformation from the .nii file. I don't know if this is what slicer does.
If you still don't manage to import your files, please post some more information so we can help you better (screen captures showing what menus you are using, what does it look like in slicer, in Brainstorn, etc).
Thank you!
Imported world electrodes but this doesn't seem to work either - the electrodes are still far away from the cortex.
I wonder whether this is because slicer uses a RAS coordinate system and SPM uses LPS. There is a suggested solution in this link but I'm not sure how to implement it...
https://www.slicer.org/wiki/Coordinate_systems
Thanks for your help!
This "world coordinates" corresponds to defined within the .nii file itself (matrix sform), not to a program or another. Can't Slicer export these coordinates within the referential of a given .nii file?
To test where the problem is, I recommend you do the following:
-
Open side-by-side the original file in MRIcron and in Brainstorm, place the cursor at the same point in the two programs and make sure that the coordinates in the title bar of MRIcron (very top of the window) matches the "World" coordinates in Brainstorm.
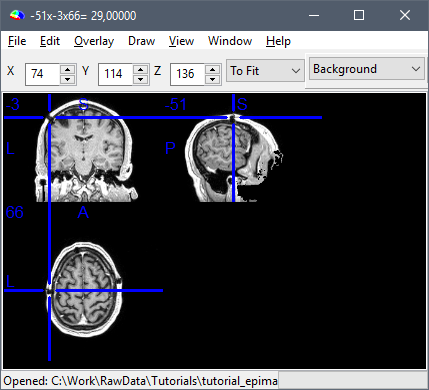
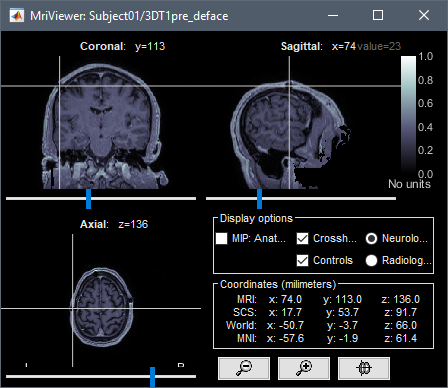
This corresponds to the "world coordinates" or "patient coordinates" associated with this .nii file (transformation matrix sform)
-
Point at the tip of one of your electrodes in either Brainstorm or MRIcron, and try to get Slicer to display (and then export) the same coordinates as what you observed previously.
If you can't find a way to view but not export these coordinates in Slicer, contact them on their forum (https://discourse.slicer.org/), reference this discussion, and post the here the reference to the thread you created.
Does it make sense?
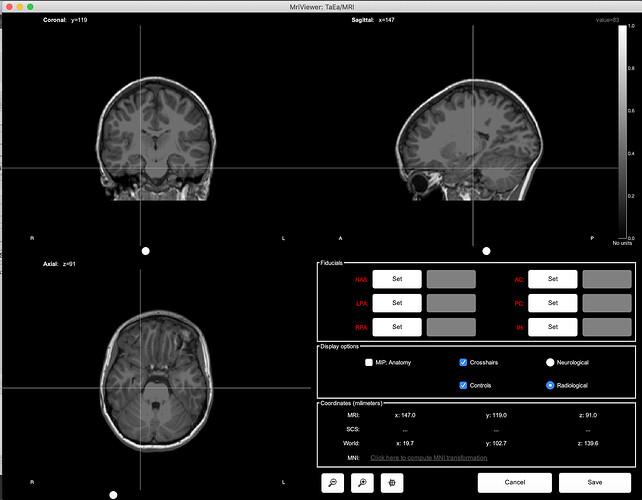
Hi Francois,
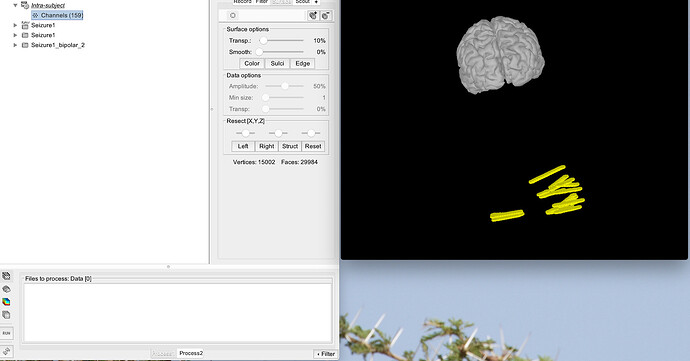
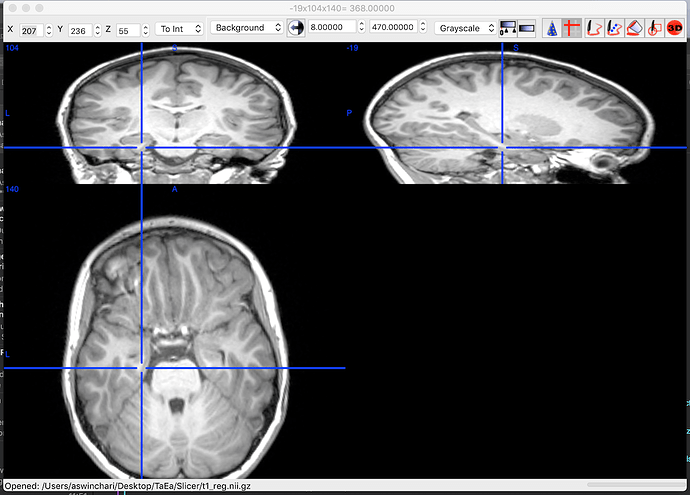
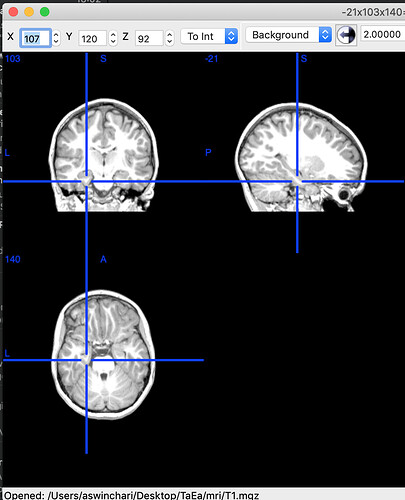
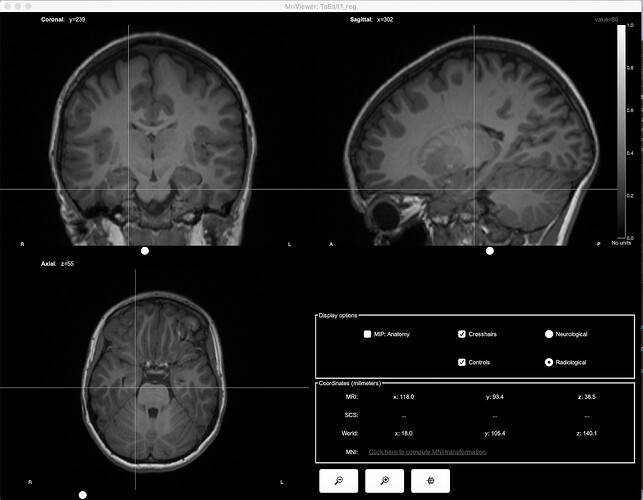
Thanks for helping with this. What you said makes sense but it seems that none of the coordinates line up. Below are 4 photos:
- MRI nit imported into MRICRON
- FreeSurfer T1 mgz file imported into MRICRON
- MRI nii imported into Brainstorm
- FreeSurfer folder imported into Brainstorm
I've picked the same location in all of them but they ALL have different coordinates!!! No idea how to resolve this.