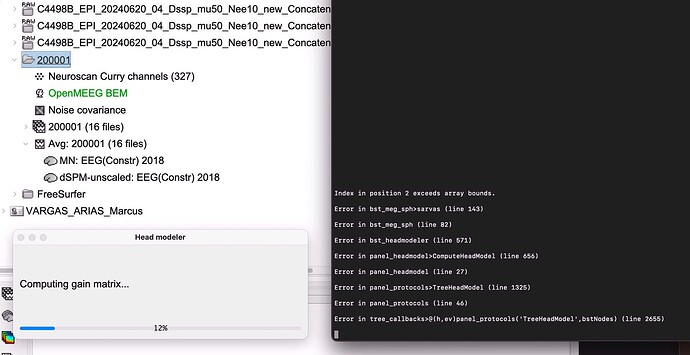
Hi,
I'm a routine Curry user and have been gradually increasing my use of BST for epilepsy source analysis. Since my Curry files have positions for both MEG gradiometers (CTF) and the EEG scalp electrodes, I have been saving my Curry MEG-EGG files as .cdt, and then loading them into BST. All goes well with the EEG, including the electrode positions, and computing a BEM head model with OpenMEEG BEM. However, with the MEG data the head model creation fails to complete—always hanging up at "computing gain matrix." This is the case whether I use overlapping sphere or OpenMEEG BEM, and whether I use either cortex or MRI volume. See attached.
Sincerely, Robert Knowlton
Hi @robertknowlton,
Would it be possible for you to share with us a simple example of the file that is causing the troubles?
Upload the file somewhere and post the download links wither in this topic or a direct message to me.
Best,
Raymundo
1 Like
Hi @robertknowlton
From the error, it looks like the MEG data format is incorrect [position, orientation, weight?].
Have you tried to visualize the MEG sensors within Brainstorm?
Can you also right-click on the channel file> file > view file content and then share the screenshot here,
It would be great to check this and then share an example of the data, as Raymundo suggested.
Thanks
Hi @robertknowlton
Apologies for lat response. I have exploring the files you sent me and these are some conclusions.
-
In Brainstorm the Curry8 importer (selecting Review raw data and file type EEG:Curry ) indeed, only supports EEG channels. As MEG sensors are read as EEG, the orientation information is lost.
-
Curry8 files with MEG sensors, do not have information about the coils, but only the centers
-
We could implement the support for Curry8 files with MEG sensors, although more example files would be needed for testing. It would great if you provide few more examples.
1 Like