Can the EEG data recorded by Nihon software be used?
Yes, files with format Nihon Kohden (.EEG) are supported. See the entire list in here:
https://neuroimage.usc.edu/brainstorm/#Supported_file_formats
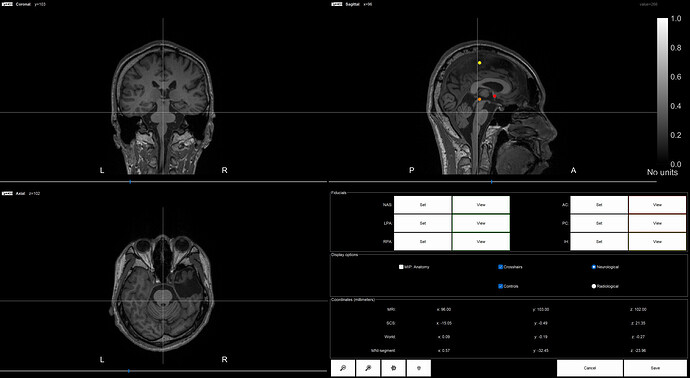
Thank you very much for your reply. Another question is what preprocessing steps should I perform before importing my preimp_T1.nii file. Is setting anatomical fiducials necessary? If so, how should I set them?
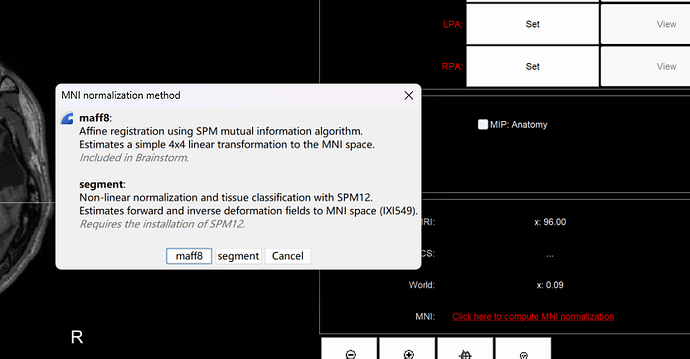
After importing preimplantation_T1, I computed MNI normalization and choose 'segment'
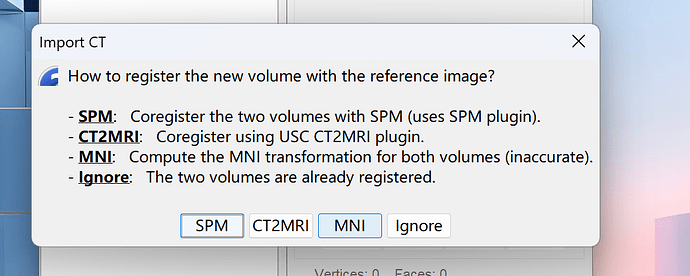
Then I imported post_CT and register it with the reference image by MNI.
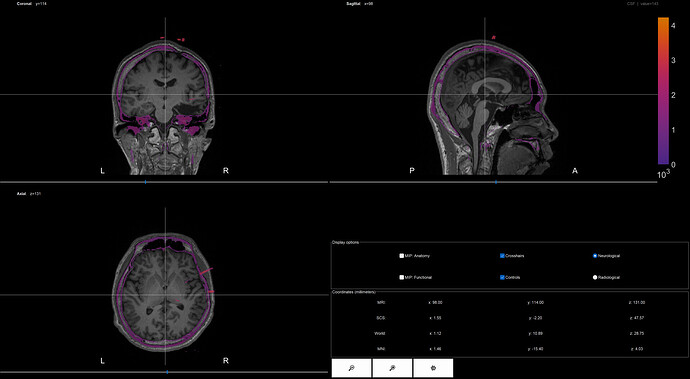
Finally, I got this
Is my operation correct?
Yes, this is needed. If you perform MNI normalization, the fiducials will be automatically set.
Either this are set during the MNI normalization, or you can set them manually (as described in this section of the Importing anatomy tutorial: https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Fiducial_points)
The co-registration of volumes looks correct. Keep in mind that co-registering using the MNI transformation may lead to inacurate results. You can find an example of the pipeline registering MRI and CT, and location of iEEG contacts in here:
https://neuroimage.usc.edu/brainstorm/Tutorials/IeegContactLocalization
- P.S.1 Please keep different topic questions on different post. This will help to other users to find useful information.
- P.S.2 If you are new Brainstorm user, we strongly recommend you to start by following the introduction tutorials with the provided dataset. This will get you familiar with Brainstorm features, processes and jargon. https://neuroimage.usc.edu/brainstorm/Tutorials
And the raw recording in Nihon Kohden database look like this
(e.g. After importing Brainstorm, previously non-existent DC channels was added.
The most important question is how to use Brainstorm to conduct group studies involving multiple patients.