Hi,
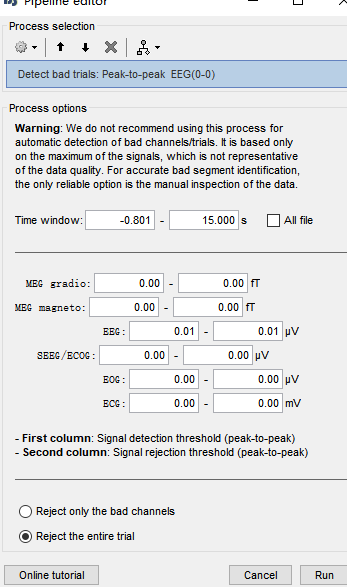
I have imported the raw data into epoches and want to detect bad trials. As Francois have suggested before, 'Artifacts-detect bad trials: peak to peak' might be a choice. I applied 4000ft as standard in my data, however, no trial was remained.
As the duration of each trial in my data was 14s, this standard might be too strict. I just want to know whether the detection could be achived by a sliding time-window instea of the total ragnge of time.
Thanks.
Not with this process.
But you can use "Events > Detect events above threshold" for this purpose.
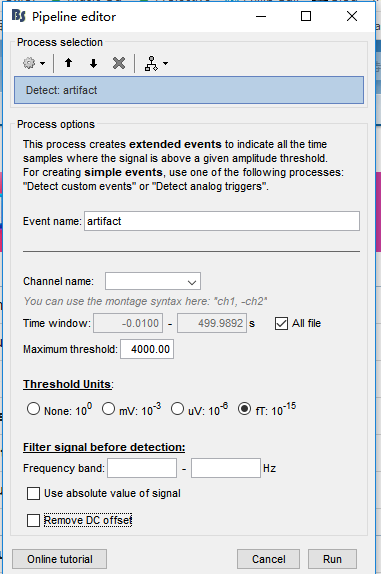
I tried "Events > Detect events above threshold" based on continuous data. I want to delet trials with ranges of values exceeding ± 4000 fT within a 1000 ms sliding time-window
at any sensor site. But I don't know how to achieve this in "Channel name".
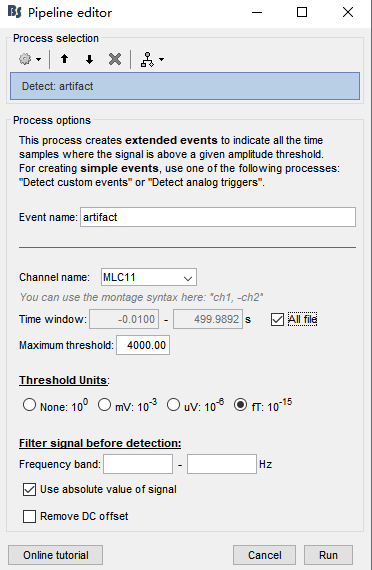
In the tutorial, the selected channel is "EEG062" (blink). I didn't record eye movement during data collection. So, I randomly chose a channel that might be contaminated by eye movement (MLC11). All other parameters are displayed in Fig 2. Am I wrong?
I want to delet trials with ranges of values exceeding ± 4000 fT within a 1000 ms sliding time-window
at any sensor site.
If you want to ignore the entire 14s trials anyway, then do the peak-to-peak detection with the first process you mentioned. I don't see the point of trying to do a detection over shorter segments.
But I don't know how to achieve this in "Channel name".
Indeed, this process works only for one channel.
You can write a script to run it in a loop over multiple channels if you need...
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting
So, I randomly chose a channel that might be contaminated by eye movement (MLC11). All other parameters are displayed in Fig 2. Am I wrong?
This is something that you have to test on your own recordings. There is no unique detection/cleaning procedure that can be applied to any type of signals, it depends on the quality of the recordings, your participant, the ambient noise, the cleaning procedures you already applied, etc...
Got it, many thanks!