Hi,
I have issues detecting bad channels and bad segments (I am quite new to preprocessing MEG data).
Briefly, at this point I have downsampled at 600 Hz and applied ICA (done through MNE python) to resting state MEG data (300 s.).
A) Bad channels
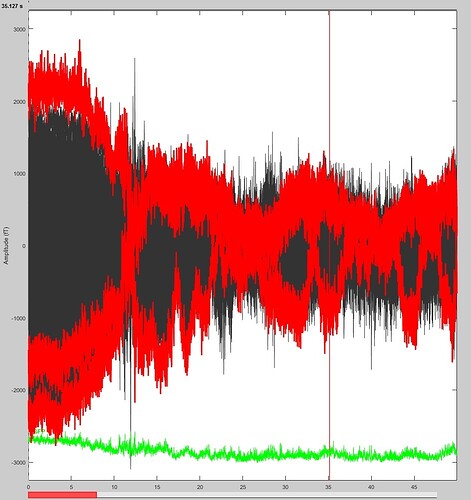
For example, in the following snapshot, can I directly infer that the channels marked in red are bad or other steps are needed in order to consider them bad channels per se? I tried to detect them trough PSD but these channels don't differ as much as shown the ones shown in your Tutorial.

B) Bad segments:
-
Procedure
To remove bad segments should I first epoched to remove bad epochs or I should go ahead and remove directly bad segments?
-
Detection of bad segments/epochs
I have problems detecting bad segments. I know there no easy answer. The bad segments illustrated in the turorial is very clear.
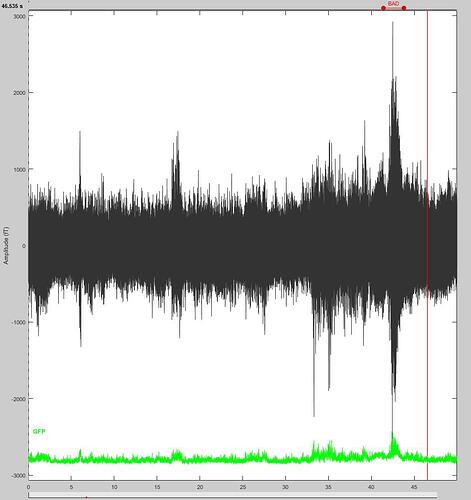
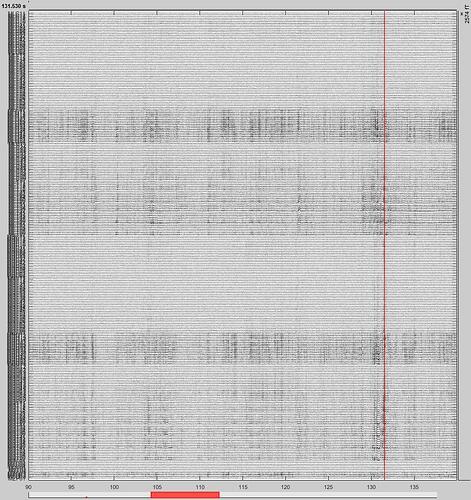
For example, in the following snapshots, would it be right to reject the segment?
Also, do you suggest manual screening for bad segments through ''butterfly'' view or all chanels separated? Because sometimes it looks ''suspicious'' only in one mode if I can put it that way. Therefore, should I consider not enough robust to justify rejection of the segment?
I also know it's important to be carefull regarding alpha band (not remove automatically high amplitude because it's not necessarilly an artefact). Therefore, should I apply a certain high pass / low pass, or proceed by thresholds to remove bad segments/epochs and bad channels?
Finally, is it suggested to proceed with your ''automatic detection'' function after looking for bad channels and bad segments ? And if so, the tutorial indicates to flag all results in each group as bad (I understand here I don't have to ask myself if the detected segments really represent artefact or not, I just simply reject them?).
Thank you in advance for your help,
Have a great day and weekend,
Véronique M.,
Étudiante au baccalauréat en psychologie
Université de Montréal
Hello,
A) Bad channels: On your screen capture, it’s very difficult to see what is happening with all the sensors overlaid. Why would you want to reject this channel from this view?
B) Bad segments:
B1. Use the procedure described in the introduction tutorials (if you haven’t read them, start by following all of them from #1 to #19).
Mark bad segments in continuous recordings, then epoch, bad trials would appear with a read dot. Then review the epochs and mark some more as bad if necessary.
http://neuroimage.usc.edu/brainstorm/Tutorials/BadSegments
http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#Review_the_individual_trials
B2. Yes, this looks bad.
- To review: the “column” mode is usually more explicit than the “butterfly”. You can review only a subset of channels at a time it the display feels too dense for you (learn how to use the keyboard shortcuts). There is no rule for all this, not everybody work in the same way.
- high pass / low pass / proceed by thresholds: it depends on your data quality, your experiment and the type of measure you are planning to compute from your recordings
- The tutorials make it clear that you should not trust any automated rejection procedure and that you should review manually all your recordings. All the detections you perform with the software should be considered as suggestions only.
After cleaning a few subjects, you’ll get used to it.
Whatever methodology you’re setting up, try to be consistent across all runs and subjects. And try not to be too aggressive on your data, removing too much is as bad as not cleaning at all.
Compute run averages and grand averages of some simple conditions to check the quality of your cleaned recordings. If you have some huge artifacts or eye movements that haven’t been removed, you’ll see them and will go back to the cleaning. It’s an iterative process, even for experts. The more experienced you are, the less iterations you need.
Good luck!
Francois
HI,
Great, thanks for the further details!
[I]Véronique M., [/I]
Étudiante au baccalauréat en psychologie
Université de Montréal