Good morning,
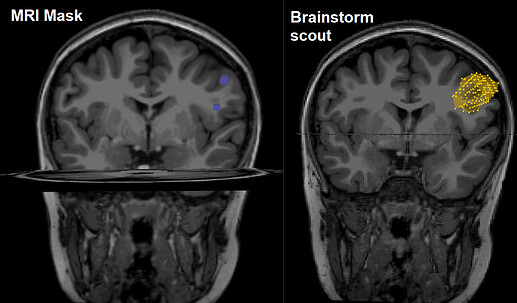
I have been trying for a few days to explore the opportunity to export a brainstorm scout to an MRI mask .nii file. I'm finding some strange anomalies in the export of some scouts (like the one I reported in the figure). Is there any reason, that I don't know, why this happens?
A second request that I have is related to the process that generates the nii mask. Is there a way to create a script and/or a function in MATLAB to generate the mask without necessarily having to import the scouts into brainstorm? I have a lot of scouts of different subjects to convert into an MRI mask and it would be very useful to create the mask automatically from Matlab interface.
Thank you for your valuable help,
Margherita
I have been trying for a few days to explore the opportunity to export a brainstorm scout to an MRI mask .nii file. I'm finding some strange anomalies in the export of some scouts (like the one I reported in the figure). Is there any reason, that I don't know, why this happens?
How do you export your mask files (.nii) and how do you expect to use them after?
Importing them back in Brainstorm seems like an unexpected thing to do (you left figure).
I double-checked that I did not introduce any bug in the export of volume scouts recently, and everything seems to work as expected. Here is what I did:
- I created a volume scout
- Exported it as
mask.nii with menu Scout > Export as MRI mask
- Exported the support MRI as
mri.nii: From the anatomy view, right-click on the first MRI volume > File > Export to file
- In MRIcron: loaded
mri.nii
- In MRIcron: menu Overlay > Add > Added
mask.nii
Both representations overlap perfectly
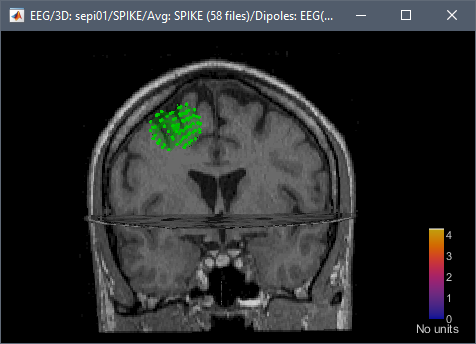
Is there a way to create a script and/or a function in MATLAB to generate the mask without necessarily having to import the scouts into brainstorm?
Do you mean export a volume scout existing in the Brainstorm database without having to display the MRI+scout and using the interface menus?
There is no function that would do that easily for you, but you can write your own Matlab script that loads the subject MRI, loads the source grid from the source file, computes the 3D interpolation and exports a new .nii file. Most calls are documented in the function panel_scout.m/ExportScoutsToMri(), and in the Scripting tutorial:
Hi Francois, thank you very much for your reply.
Actually, my left figure was done in DSI studio after I had imported the .nii file mask of the scout shown on the right. I need that specif ROI to track fibers from DTI. As for the export of the mask, I followed exactly the steps you wrote. This is why the difference between the two volumes seemed so strange to me.
However yesterday after a few attempts I understood the problem. I created a mask by selecting two partially overlapping scouts, mistakenly thinking that they were both considered correctly. Instead in the creation of the mask, the largest scout is correctly rebuilt but the second is "cut" and only the parts not included in the other scout are kept. My mistake sorry, I have misused the mask export.
As for the second question, thank you, it was exactly what I was looking for and I will try to implement it.
If two scouts overlap, they can't be both represented in the volume.
The export of multiple ROIs is done interatively: The voxels included in scout #1 get value 1, then the voxels included in scout #2 get value 2, etc.
Therefore scout 1 would appear as cut, if processed alone.