Dear Francois,
I am using someone else's dataset, who did not use Brainstorm to do their headshape digitization, so the file is in .pol format and I don't have a .pos file.
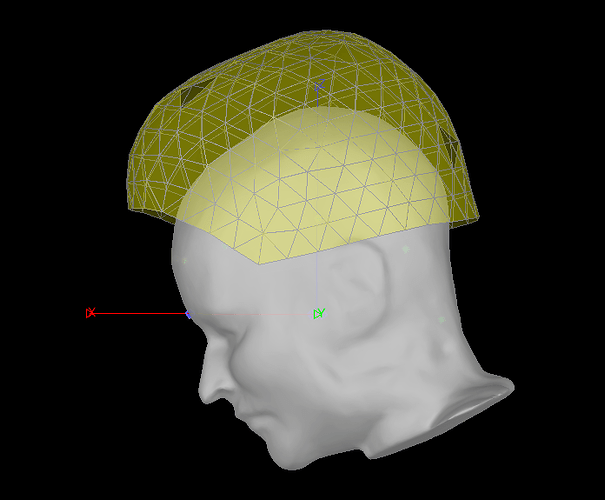
The first three points digitized were the HPI coils, and afterwards they did multiple trajectories over the head in an anterior-to-posterior direction and side to size, so points all over the head and some on the brow and nose that I suggested. They sent me a 3D matlab figure that looks entirely normal to our standard McGill digitization strategy.
However, I'm having trouble using it.
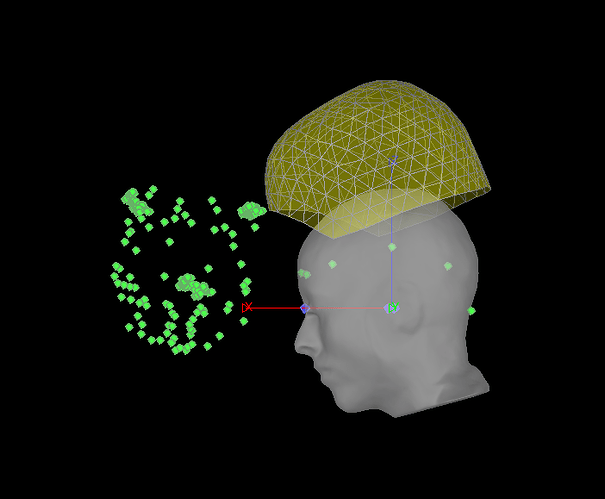
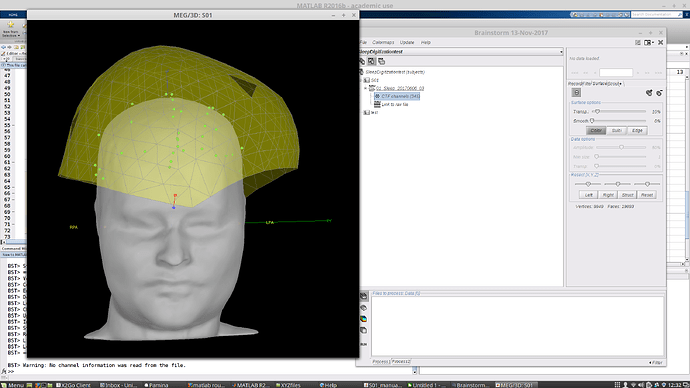
When I first read in the raw .ds folder, the surface comes up with some points already on it. These have EEG labels, though they are not all EEG and are in the wrong places, so I don't intend to use them and they aren't meaningful. At this stage it looks like this:
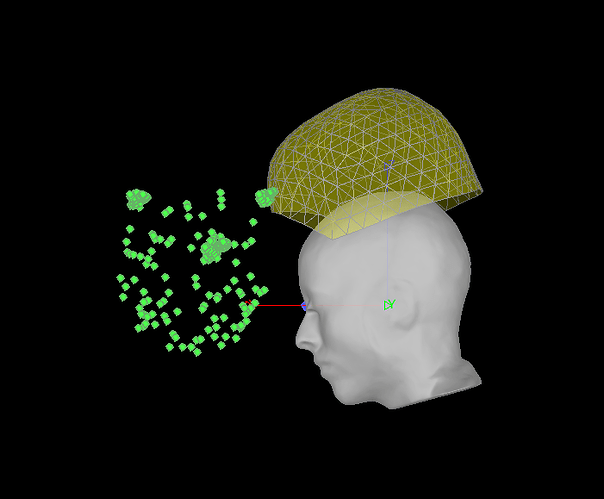
If I right-click on the file > Digitized head points > Add points > and navigate to my .pol file, it tells me "315 new head points added. 315 duplicate points (ignored). Total 325 points" and adds a shell of points in a funny orientation. I also notice that there are three clusters of large numbers of points that are probably at the HPI coils, but I don't think that these were digitized by my colleague as they don't make much sense and were not present in that matlab 3D view I mentioned above. Now it looks like this:
If I attempt to use these head points to refine registration, nothing changes and it says: " The optimization failed finding a better fit"
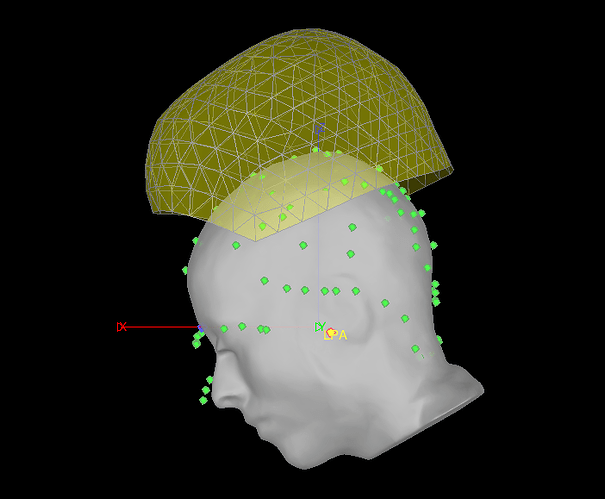
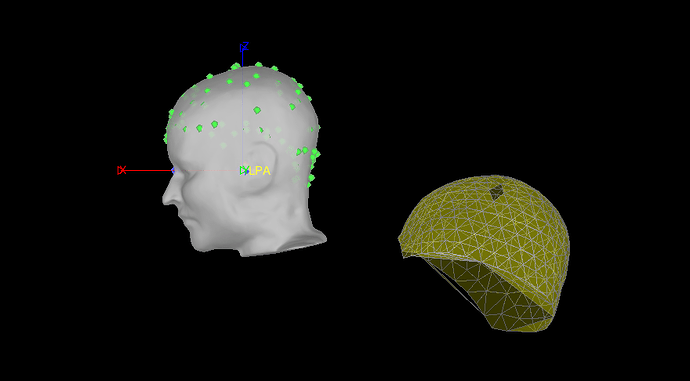
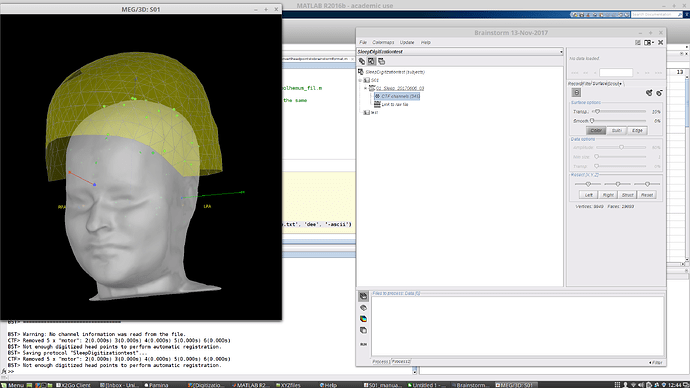
Since I believe the EEG points are useless, I went to Digitized head points > Remove all points. I then tried to reload the .pol file. Now the EEG points are gone and it looks like this:
If I refine using head points now, I still get no change and a warning that a better fit was not found.
I looked at some forum topics and found the suggestion of reformatting the .pol data in the same format as the .pos files. However, looking at a past (successfully used) .pos file as an example, it' s not clear what's what. In the good file, I have abut 430 lines each numbered sequentially, and then 3 coordinates, and at the end there's the nasion, LPA and RPA twice. 430 is more than the number of points that were collected, I believe.
In the new, problematic .pol file, I have about 315 lines, and the first column repeats the sequence 01-04. And then there are 6 instead of 3 columns of presumably coordinates. Every number 01 has an extra column that has a 1 in it, but in one case a 0. Here is the first bit:
01 27.62 26.77 7.56-178.20 4.48 28.43 1
02 25.98 34.09 9.83 130.25 23.66 -85.92
03 19.67 34.41 1.50 36.42 -45.01 58.85
04 14.06 31.64 9.70 69.29 64.96 93.85
01 14.11 24.35 8.31 5.95 17.98 88.46 0
02 24.92 33.92 9.94 131.68 22.87 -84.48
03 18.54 34.14 1.69 36.90 -44.69 60.84
04 13.23 31.28 10.04 72.73 65.67 96.30
01 18.89 33.41 4.10 -74.23 12.51 79.10 1
02 25.18 33.93 9.94 130.72 23.13 -84.74
03 18.85 34.13 1.62 36.74 -44.48 60.09
04 13.53 31.31 9.96 70.63 65.26 94.67
(...)
So now I' m out of ideas about how to use or convert this .pol file! Do you have any suggestions?
Thanks,
Emily