Hi there!
After ft_dipolefitting the Avg (epoched data), I get the fitting of a dipole and I can see it in MRI and cortex GUI which is where this dipole is.
How can I make sure to correctly report the calculated dipole points that I see in the MRI or cortex as a region of interest based on the desikan killiany atlas or any other? FYI, mostly the dipoles are around the thalamus, fornix and frontal part. I would appreciate any hint!
Cordially
Amir
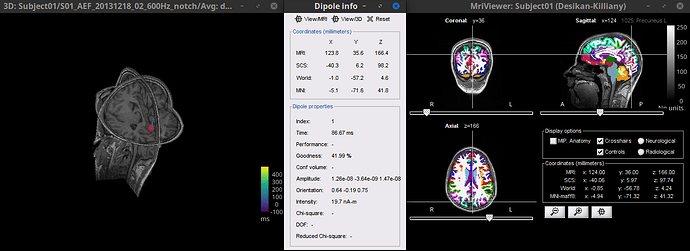
Double-click the dipole file (![]() ), it will open a figure showing the dipoles, click on the dipole of interest and a window showing their coordinates will appear. Now click on View MRI to open the MRI viewer, and in this figure it is possible to overlay anatomical atlases, the legend at the top-right corner will indicate if the dipole position corresponds to a parcellation of the selected atlas.
), it will open a figure showing the dipoles, click on the dipole of interest and a window showing their coordinates will appear. Now click on View MRI to open the MRI viewer, and in this figure it is possible to overlay anatomical atlases, the legend at the top-right corner will indicate if the dipole position corresponds to a parcellation of the selected atlas.
This figure shows a dipole (left), its coordinates (center) and the parcellation where it falls in the Desikan-Killiany atlas (right)
1 Like