Hi,
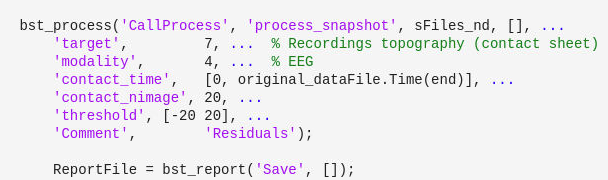
I'm using the approach in the image to generate and later save contact sheets of EEG topography. How could I include the sensor labels on the topography in this code? I am unable to find this from the original codes on github.
Thank you!

Hi,
I'm using the approach in the image to generate and later save contact sheets of EEG topography. How could I include the sensor labels on the topography in this code? I am unable to find this from the original codes on github.
Thank you!
Hi @vidushani, the process snapshot offers only the basic settings for such plots.
More advance customization can be done scripting. The following snippet can replace the call to process process_snapshot:
% Parameters
modality = 'EEG';
contact_nimage = 20;
contact_time = [0, original_dataFile.Time(end)];
displayMarkers = 1;
displayLabels = 1;
% Display topography
hFig = view_topography(sFiles{1}, modality, '2DSensorCap');
set(hFig, 'Position', [200, 200, 200, 220]); % Default position for contactsheets
bst_colormaps('SetDisplayColorbar', modality, 0); % Do not include colorbar
% Show sensor markers and labels
figure_3d('ViewSensors', hFig, displayMarkers, displayLabels, []); % Add Markers and Labels
% Contactsheet
hFig = view_contactsheet(hFig, 'time', 'fig', [], contact_nimage, contact_time);
% Add to report
bst_report('Snapshot', hFig, sFiles{1}, '', []);
% Close figure
close(hFig);
Thank you Raymundo! That was very clear.