Hello,
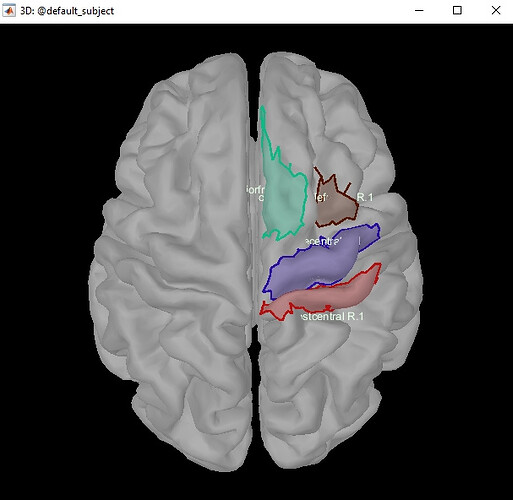
I made a subdivision of the DK atlas on the default anatomy. Saved it as a .mat file. Looks like so.
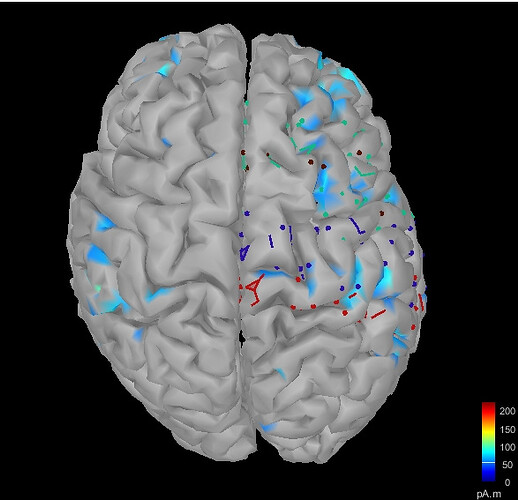
For sources computed from patient anatomy, when i load this subdivided atlas .mat file, it looks like this. What am i doing wrong? The default DK atlas looks fine, but not my saved file.
(Im running a PLV code separately and basically i want the tess_cortex_pial_low.mat file from the projected file into the normalised atlas. I dont know if the sources projected to the normalised atlas are correct or not)
Regards
Gajanan
Brainstorm - update feb2018
Matlab 2017
Windows pc
Hello,
If I understand correctly, you create the ROIs on the anatomy template, save them as .mat files and try to load them in a subject.
This cannot work because the cortex meshes for the template and the subject are different, the vertices do not match point-to-point, and the scouts are simply lists of vertex indices.
If you have processed the subject MRI, you have access to an interpolation matrix with which you can project source maps or regions of interests (http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects).
When displaying the ROIs you designed, go to Surface tab, menu Scout > Project to > select the appropriate patient’s surface.
Cheers,
Francois
Crystal. Thank you Francois!
Was missing this step --> [When displaying the ROIs you designed, go to Surface tab, menu Scout > Project to > select the appropriate patient’s surface.]. pial_low has that info now.
Cheers,
Gajanan
Hi. I had the same issue and I tried to project the ROIs I selected. It worked fine for the default anatomy. But when I tried to project to other individual MRIs, I had the the same problem as if I were saving them as .mat. Is there a way to create an customized atlas with selected ROIs (including subdivided scouts) and make it be recognized like it does with the DK atlas?