Dear Experts:
Good morning!
We have already preprocessed the EEG data (ICA, epochs, reference, bad channel removal, etc.) in the EEGLAB.
However, we need to use Brainstorm to analyze brain connectivity further. We have imported the preprocessed EEG data into the Brainstorm using 'import EEG/MEG.' Do we need to preprocess everything again in the Brainstorm (i.e., channel location, ICA removal, bad channels detection) for connectivity analysis?
Thank you so much
Zhepeng Rui
If you have imported pre-processed data for which the modifications were applied to the EEG data, you can use it directly in Brainstorm.
If you have imported the original raw data, you may need to clean it again.
Thank you! Francois!
You are so professional! I saw you replied to so many helpful comments from the public!
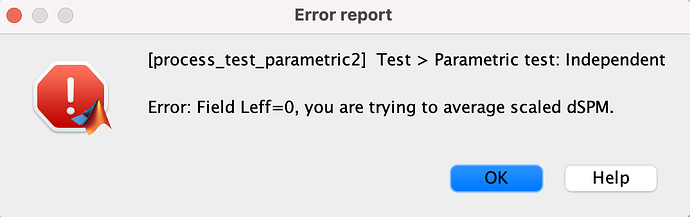
By the way, we met one problem on statistics analysis towards scaling or unscaled dSPM volume source estimation on parametric t-test. This method does not seem to allow scaled average dSPM analysis, but it works for unscaled versions. The permutation t-test also worked for scaled dSPM analysis.
So, does that mean Parametric test: independent is not working for scaled dSPM analysis?
Thank you
Zhepeng Rui
Indeed, as stated in the introduction tutorials:
" Ajusting an averaged dSPM file by this sqrt(N) factor is still possible manually, eg. in order to visualize cortical maps that can be interpreted as Z values. Select the average dSPM files in Process1 and run process "Sources > Scale averaged dSPM". This should be used only for visualization and interpretation, scaled dSPM should never be averaged or used for any other statistical analysis."
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Averaging_normalized_values
Thank you, Francois! Thank you for such quick response! Therefore, is un-scaled dSPM qualified for statistical analysis based on publication standards?
Thank you!
Here is the final question for this part. We have known that the scaled dSPM is used for visualization and interpretation. If we used the unscaled dSPM to do the statistical analysis, do we need to show up the scaled dSPM brain map or unscaled dSPM brain map for the publication standards?
Best regards!
Zhepeng Rui
If we used the unscaled dSPM to do the statistical analysis, do we need to show up the scaled dSPM brain map or unscaled dSPM brain map for the publication standards?
You can run the group statistics on unscaled dSPM values, and show individual averages as scaled values.
Thank you! You are so accommodating and kind!
Best regards!
Zhepeng Rui