Dear Brainstorm team,
I am trying to generate FEM headmodels with DUNEuro in Brainstorm :
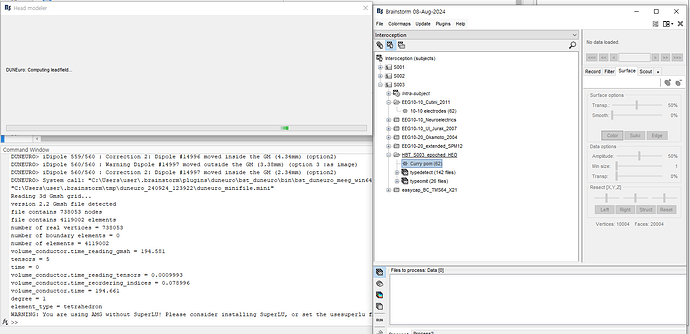
I tested it on 20 subjects, and it works correctly for 17 of them. However, for 3 subjects, the process gets stuck during the "Computing leadfield" step.
What happens is that after the warning about SuperLU, there is no error message, but the computation keeps running for hours without progressing (unlike the other subjects, where the process usually takes only a few hours and finishes successfully).
I don’t think this is related to the computational capacity of my computer, since it works fine for the majority of subjects. The three problematic subjects do not seem different from the others.
-
I used the option "force in grey matter" (and also tried resizing/reducing the cortical surface to make it closer to the mesh).
-
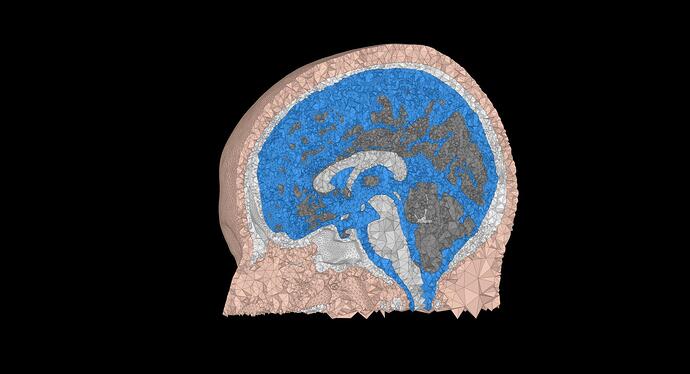
I am using a cortical surface from FreeSurfer and a FEM mesh generated from SimNIBS (because I want to keep the cerebellum).
-
When there was a mismatch between cortex and mesh (vertices "out of grey matter"), I used to get explicit error messages but here I do not get any errors, just an infinite loop with no update in the Matlab command window.
Do you have any suggestions on what I could check for these specific subjects? Are there recommended verification steps to identify why the FEM computation would hang without producing an error?
Any help or advice would be very much appreciated.
Thank you in advance!
Best regards,
Candice
Code :
% Input files
sFiles_hm = filteredList(filter);
% Start a new report
bst_report('Start', sFiles_hm);
% Process: Compute head model
sFiles_hm = bst_process('CallProcess', 'process_headmodel', sFiles_hm, , ...
'Comment', '', ...
'sourcespace', 1, ... % Cortex surface
'meg', 1, ... %
'eeg', 4, ... % DUNEuro FEM
'ecog', 1, ... %
'seeg', 1, ... %
'duneuro', struct(...
'FemCond', [0.14, 0.33, 1.79, 0.008, 0.43], ...
'FemSelect', [1, 1, 1, 1, 1], ...
'UseTensor', 0, ...
'Isotropic', 1, ...
'SrcShrink', 0, ...
'SrcForceInGM', 1, ...
'FemType', 'fitted', ...
'SolverType', 'cg', ...
'GeometryAdapted', 0, ...
'Tolerance', 1e-08, ...
'ElecType', 'normal', ...
'MegIntorderadd', 0, ...
'MegType', 'physical', ...
'SolvSolverType', 'cg', ...
'SolvPrecond', 'amg', ...
'SolvSmootherType', 'ssor', ...
'SolvIntorderadd', 0, ...
'DgSmootherType', 'ssor', ...
'DgScheme', 'sipg', ...
'DgPenalty', 20, ...
'DgEdgeNormType', 'houston', ...
'DgWeights', 1, ...
'DgReduction', 1, ...
'SolPostProcess', 1, ...
'SolSubstractMean', 0, ...
'SolSolverReduction', 1e-10, ...
'SrcModel', 'venant', ...
'SrcIntorderadd', 0, ...
'SrcIntorderadd_lb', 2, ...
'SrcNbMoments', 3, ...
'SrcRefLen', 20, ...
'SrcWeightExp', 1, ...
'SrcRelaxFactor', 6, ...
'SrcMixedMoments', 1, ...
'SrcRestrict', 1, ...
'SrcInit', 'closest_vertex', ...
'BstSaveTransfer', 0, ...
'BstEegTransferFile', 'eeg_transfer.dat', ...
'BstMegTransferFile', 'meg_transfer.dat', ...
'BstEegLfFile', 'eeg_lf.dat', ...
'BstMegLfFile', 'meg_lf.dat', ...
'UseIntegrationPoint', 1, ...
'EnableCacheMemory', 0, ...
'MegPerBlockOfSensor', 0), ...
'channelfile', '');
% Save and display report
ReportFile = bst_report('Save', sFiles_hm);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
% bst_report('Email', ReportFile, username, to, subject, isFullReport);