Hi Brainstorm community.
I have warped electrode locations from MNI to all my subjects using non-linear registration of T1 to template.
My electrode locations are in the subject T1 space in mm, and I checked them in freeview.
The T1 have been processed using freesurfer and imported in brainstorm interface.
When loading the electrodes location as text file XYZ, it suggests me to scale it (0.1 to cm) but the cap is not at all aligned (rotated and translated, scale seems OK) to the surface in Brainstorm.
What is the coordinated system for this XYZ import?
Is there a way to convert the mm in MRI space to a format that brainstorm would accept?
Thanks for you help.
Hello,
The coordinate system Brainstorm uses for the surfaces and the electrodes positions is based on the anatomical fiducials NAS/LPA/RPA (SCS):
http://neuroimage.usc.edu/brainstorm/CoordinateSystems#Subject_Coordinate_System_.28SCS_.2F_CTF.29
The easier way to have your positions converted automatically to this internal coordinates system is to add these three points NAS/LPA/RPA in your file.
Mark them in freeview exactly where you place them when you import the subject anatomy in Brainstorm.
When importing the positions in Brainstorm, it would use them to compute the transformation MRI=>SCS.
Does this help?
Francois
Hi François.
Thanks for your quick answer.
I tried adding NAS/LPA/RPA but it does not help. Also the fact that we can import also without electrode name shows that it does necessarily requires the landmark to import.
Looking at the code (in_channel_ascii.m), it seems that first the loaded coordinates are rescaled by .01, then it also ask for another factor if it does not seems right and apply this second factor.
I tried to use cs_convert from the mri location (xyz in mm) to scs coordinates which seems the format required for Channels.Loc field but it does not work either.
I also tried to apply sMRI.SCS transformation manually [SCS.R SCS.T ; 0 0 0 1] to the mm MRI coordinates and playing with the scaling but there still exists a translation.
It seems that MRI coordinates space of brainstorm is very different from the standard nifti definition and that no clear transformation exists from one to the other and to the SCS space required for electrode locations.
Do you have an idea of a way to convert? It does not need to be using the interface.
Thanks.
The Brainstorm MRI coordinates are the voxel coordinates, which can be very different from the “world coordinates” you can get from another volume viewer.
If you have the option of adding the NAS/LPA/RPA in the same coordinates system as the electrodes, you should be able to get your electrodes registered automatically.
If you can’t get this to work, there is something wrong with the files in which you save your coordinates.
Can you post your file here? (or send it to me by email: click on my username on this forum)
Thanks
Francois
Ok, looking at the code it seems that MRI coordinates = voxel coordinates scaled by voxel size (with a specific origin voxel).
my file looks like:
Fp1 59.1643 29.2384 62.022
Fp2 57.2951 -34.8589 61.4726
F3 10.4206 47.9692 82.6552
F4 6.72031 -50.3491 81.9799
C3 -51.5151 62.0092 68.3701
C4 -56.1684 -61.1301 69.2636
P3 -95.8378 48.2995 26.7391
P4 -100.376 -43.3072 26.2099
O1 -98.0094 29.0075 -19.4209
O2 -100.709 -23.5801 -20.1083
F7 22.5117 65.7616 35.8602
F8 17.184 -68.9918 35.6902
T7 -22.4806 77.4168 11.38
T8 -28.4466 -76.9808 11.3372
P7 -67.4291 66.7957 -7.78926
P8 -72.1761 -63.0349 -7.99957
Fz 4.62001 -1.52606 105.73
Cz -68.7455 0.751849 99.7921
Pz -113.292 2.69047 42.1627
Oz -103.608 2.80707 -21.5944
FC1 -28.3727 32.4515 98.7306
FC2 -30.2513 -33.0324 97.8858
CP1 -91.7333 34.0176 70.9526
CP2 -94.7215 -30.8455 70.9746
FC5 -11.1549 71.4476 51.1421
FC6 -16.6774 -72.8516 51.4162
CP5 -60.372 72.2138 26.5105
CP6 -66.101 -70.6925 26.6808
POZ -113.963 3.41594 10.443
F1 6.77404 25.6051 99.2038
F2 4.93609 -28.9441 98.8446
C1 -62.5577 37.1342 89.5414
C2 -65.1174 -35.5864 89.6036
P1 -107.777 25.8633 37.998
P2 -110.871 -20.761 38.2991
AF3 40.8623 33.5101 82.3553
AF4 38.1877 -38.6994 82.3331
FC3 -19.2193 56.7173 79.184
FC4 -23.624 -57.2581 78.7234
CP3 -78.5769 59.4772 51.1701
CP4 -83.882 -57.4865 51.6477
PO3 -104.768 33.4699 3.05256
PO4 -107.894 -27.5944 2.52779
F5 15.394 61.4752 60.8796
F6 10.6538 -64.1453 60.5592
C5 -36.4174 75.4617 39.1089
C6 -42.0971 -75.1287 39.7369
P5 -80.7482 61.022 11.0752
P6 -85.3889 -57.3698 10.5784
AF7 42.6585 52.1908 51.0912
AF8 38.9943 -56.1435 50.6475
FT7 -0.0740746 73.8895 22.0902
FT8 -5.93296 -75.0909 21.8234
TP7 -45.031 74.6915 0.858351
TP8 -50.9612 -73.5108 0.193501
PO7 -85.8359 50.7931 -14.7517
PO8 -89.9109 -46.4618 -15.2286
FT9 8.32271 70.3891 -2.19418
FT10 1.99999 76 -21
FPZ 5.77106 -69.7953 -1.68878
CPZ -6 -79 -25
FCz 61.1622 -3.12503 67.3695
NAS 125 -7.99999 -7
RPA -97.227 2.03418 78.1387
LPA -30.6882 -0.53913 108.428
This is what it looks like after import:
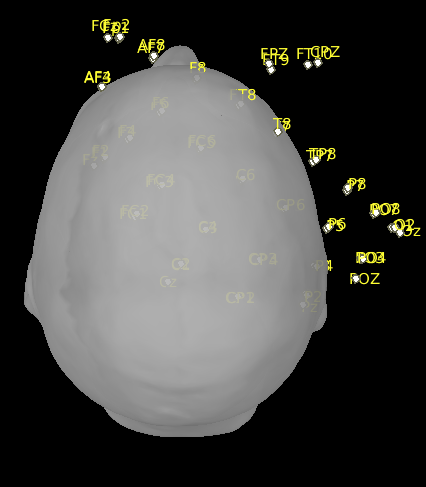
I have been reading the code of import_channel.m but cannot find any link to an automatic registration triggered after import.
Do you mean using "refine using head points"? I also tried that and it just align but really weirdly, here it what it looks like:
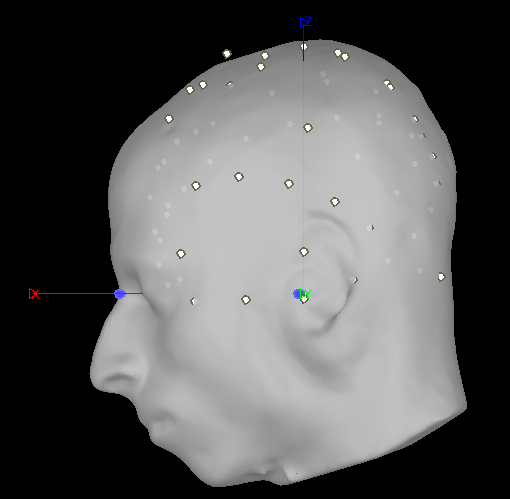
Am I doing something wrong?
Thanks for your help.
Basile
Hi Basile,
You don’t have anything to do: when the three points NAS/LPA/RPA are present in a file, it is automatically converted to the Brainstorm subject coordinate system (SCS), based on these three points.
You result is wrong just because the position your LPA and RPA points is wrong (just rename them to see them after importation):
- LPA is at the position of FCZ
- RPA is at the position of PCZ
Check your export procedure…
Francois
Thanks, I didn’t check what freeview reexported from my points file, and in fact in just transforms all the points that I loaded without any reasons (even reimporting in freeview is no longer aligned to MRI), plus the points that I added manually were different too, very weird…
Sorry for bothering you with this issue and thanks again for you kind help.
Basile

