Hello,
I have a EEG protocol with 22 subjects, and 27 conditions.
For each subject, I would like to compute the noise covariance matrix from one condition (which contain a section define as 'baseline'). Also, I apply this noise covariance matrix to all the 27 condition of my subject.
Then, I reproduce this process for my 22 subjects.
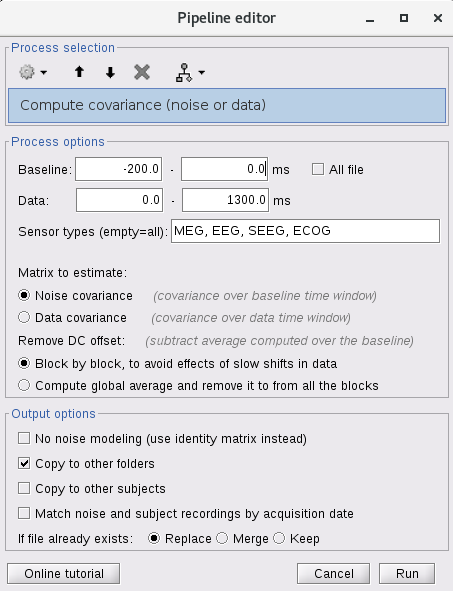
I 'semi'-automatized it with Process1 > Sources > Compute noise covariance > Process option: edition of the baseline time-window and use the following parameter:
However, my question is: it is possible to fully automatized this process? In other word, to perform the same for all subject at the same time, and not drag/drop them one by one.
Indeed, when I tried to add all participants in Process1 section, BST compute the noise covariance matrix for the first of the list and apply this matrix to all participants.
Thank you!