Hi everyone!
I have preprocessed EEG data in .set format (from eeglab).There is no individual fMRI data. As I understand it, if I want to do source localization, I need to warp the template or to project electrodes on the surface.
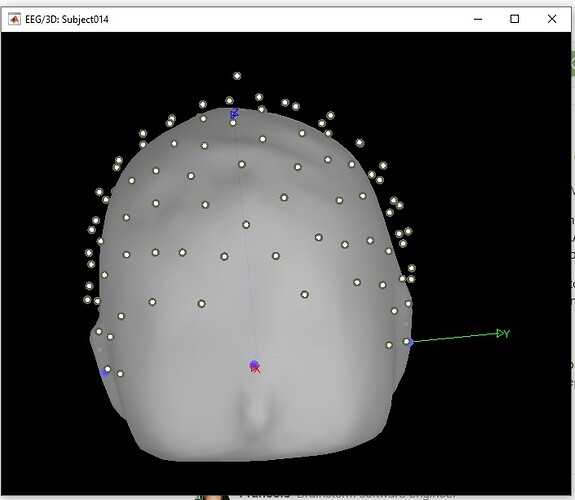
But in my set files there are no fiducial points (only 128 standart channels). I have coordinates of fiducials but then we preprocess the data in eeglab, we don`t add them. So should I add these 3 points now in the BST and how can I do that?
Kind regards,
Kate
If these are standard 10-10 positions added by EEGLAB: you can discard them and use the positions already adjusted to the Brainstorm MNI template (ICBM152).
Right-click on the the channel file > Add EEG positions > ICBM152 > ...
If these are digitized positions: import the positions again from Brainstorm instead of using the EEGLAB version, they might contain registration information that EEGLAB skipped.
Right-click on the the channel file > Add EEG positions > Import from file.
If this is a non-standard cap (or simply not available in Brainstorm yet) for which you have standard positions in EEGLAB only: then you have no other choice than adjusting the positions of the electrodes manually on the head surface.
Right-click on the channel file > MRI registration > Edit.
Additional help from processing EEG in Brainstorm in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Prepare_the_channel_file
PS: For help with source localization on EEGLAB files, you can search the forum, there are many people who asked about that previously
Thank you for the answer! I will try import the positions again.
Francois, I'm sorry to bother you again.
I followed your advice to import the EEG positions again from Brainstorm instead of using the EEGLAB version.
But it seems like formats that I have don't suit BST. None of that was read properly.
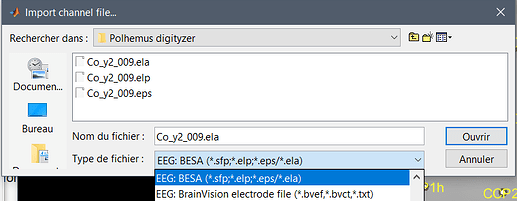
I have ela,elp,dat, 3dd,eps formats, if you want to see it's here: https://yadi.sk/d/zN2BU6xPMg68Bg
Then I import .set file, BTS suggests me scaling factor '10', and I think it looks pretty normal, just a litlle far from head surface. Can I just use this and project it on the surface (by MRI registration --> edit)?
As I cant use my formats of EEG positions, I cant add fiducials.
I manually created .pos file with 3 points (from .eps file).
It looks like :
1 LPA -6.127171 -0.314485 0.168297
2 NAS -0.117323 9.289759 -0.004542
3 RPA 6.639113 -0.213330 0.000657
then digityzed head points -->add point'.
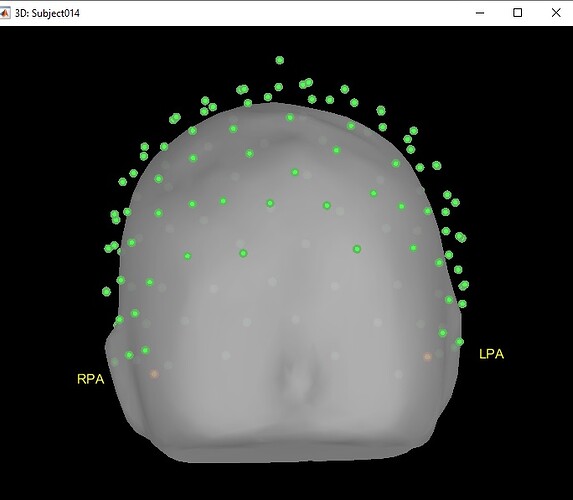
But it read only 2 points (lpa, rpa) and they are inside head.
Can you help me with this problems?
thanks
Kate
If you select the file format "EEG: BESA", it reads correctly at least the .ela and .eps.
With the format "EEG: Neuroscan", it reads correctly the .dat.
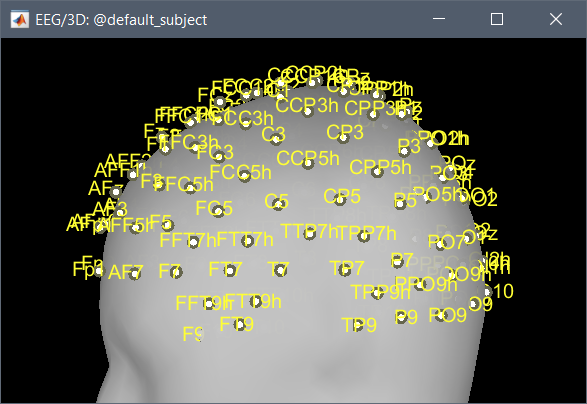
Can I just use this and project it on the surface (by MRI registration --> edit)?
You need to adjust manually the registration:
- Right-click on the channel file > MRI registration > Edit
- Rotate/translate/resize to get as close as possible to the head surface. Read the tooltips of the buttons in the toolbar for help
- Finally project the electrodes on the head surface
I can't add fiducials
it read only 2 points (lpa, rpa) and they are inside head.
It doesn't make much sense to add these points manually after importing the channel file.
If they are present in the file you import as your "channel file", the fiducials NAS/LPA/RPA are used automatically to convert the positions in the channel file. And then they are removed from the list of channels.
If you want to add the fiducial positions, you need to edit the text file with the 3D position of the electrodes and add them in there. This is possible only with a file format where the electrodes are named.
There are NAS/LPA/RPA points defined in the .dat, but their position do not make sense, relatively to the EEG channels...
The simplest would be for you to adjust this registration manually...