If you select the file format "EEG: BESA", it reads correctly at least the .ela and .eps.
With the format "EEG: Neuroscan", it reads correctly the .dat.
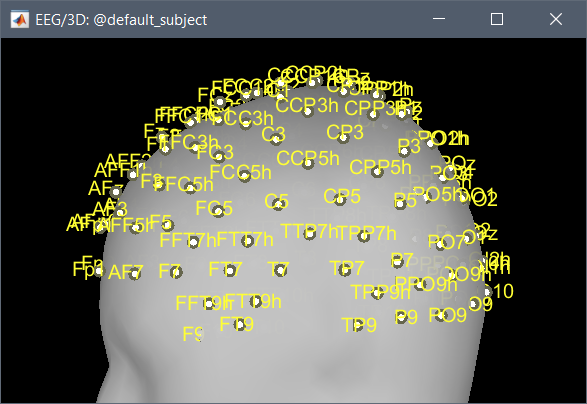
Can I just use this and project it on the surface (by MRI registration --> edit)?
You need to adjust manually the registration:
- Right-click on the channel file > MRI registration > Edit
- Rotate/translate/resize to get as close as possible to the head surface. Read the tooltips of the buttons in the toolbar for help
- Finally project the electrodes on the head surface
I can't add fiducials
it read only 2 points (lpa, rpa) and they are inside head.
It doesn't make much sense to add these points manually after importing the channel file.
If they are present in the file you import as your "channel file", the fiducials NAS/LPA/RPA are used automatically to convert the positions in the channel file. And then they are removed from the list of channels.
If you want to add the fiducial positions, you need to edit the text file with the 3D position of the electrodes and add them in there. This is possible only with a file format where the electrodes are named.
There are NAS/LPA/RPA points defined in the .dat, but their position do not make sense, relatively to the EEG channels...
The simplest would be for you to adjust this registration manually...