Hello All,
I imported my cleaned, averaged ERP data from EEGlab to Brainstorm in order to further run some source localization analysis. My data was already average referenced prior to import during pre-processing, which means that it has an additional channel for Cz, turning 64-channel ERP data to 65 channel ERP data.
(Raw data was recorded with EGI GSN_Hydrocel_64 channel -vertex reference).
I have two questions:
- What are coordinates for Cz that aligns with the head model ICBM152?
I am using the default head model ICBM152, and when I use the default channel locations (Add EEG Positions > ICBM152>EGI>GSN_HydroCel_64_E1, it automatically detects the correct channel locations and adjust it according to the head model; but with a missing electrode for Cz. I tried to add the Cz to the file by 1)manually adding an electrode via GUI>MRI Registration>edit, 2) manually adding a line for "channel 65" in the original mat file "channel_GSN_HydroCel_64_E1.mat" which is located under /Users/ddc/Documents/brainstorm3/defaults/eeg/ICBM152).
However, since I don't know the exact coordinates for Cz for the specific head model, my additions are approximate, which is probably not a very good solution. I was wondering if there was already coordinates established for Cz (for the ICBM152 head model) ? I do have an external .sfp file that includes Cz as the 65th channel (works in EEGlab) but it does not fit the ICBM152 head model when I import it.
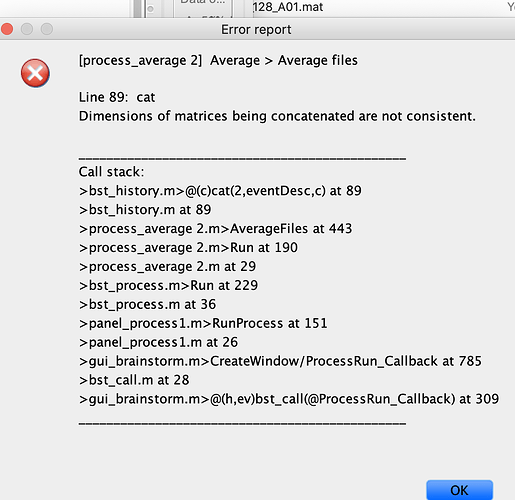
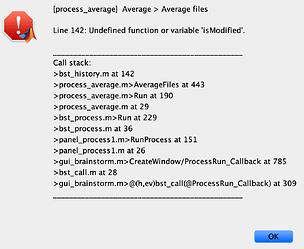
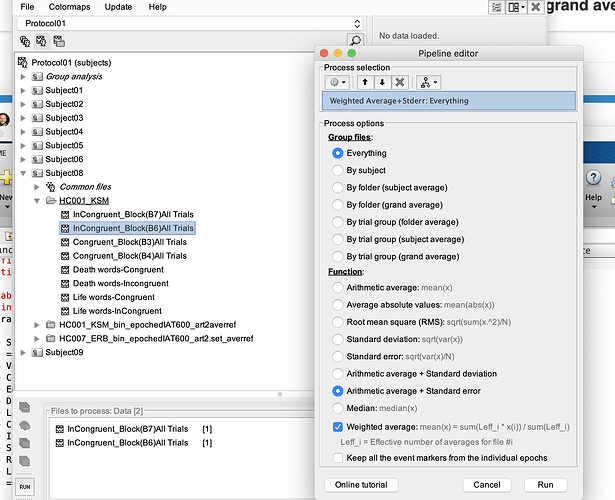
- Another problem I am having is when I try to average ERPs across subjects for (grand average). I thought it may be related to channel locations so I am including this in the same thread. This is the error I get (any of the averaging options would give me the same error (by trial/subject/folder):
Unfortunately I am not savvy enough to track the error to its origin but I thought this might be due to differences in channel arrays (vertical vs horizontal), but I am not sure if this is the case or there is another problem. I get the same error whether or not I manually edit the channel location file to have the Cz included as the 65th electrode with [1 x 65] structure.
My ERP data has the following structure (one of the ERP categories as an example):
|- F: [65x800 double]
|- Std: [65x800 double]
|- Comment: 'InCongruent_Block(B7)All Trials '
|- ChannelFlag: [65x1 double]
|- Time: [1x800 double]
|- DataType: 'recordings'
|- Device: 'ERPLab'
|- nAvg: 64
|- Leff: 1
|- Events: [0x0 struct]
|- ColormapType:
|- DisplayUnits:
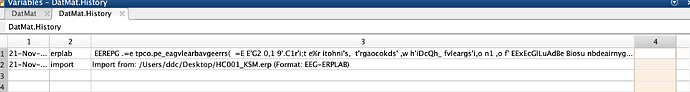
|- History: {2x3 cell}
.
I have Matlab 2016a, and MacOS(Catalina)
Thanks so much for your help!
Deniz