Hi,
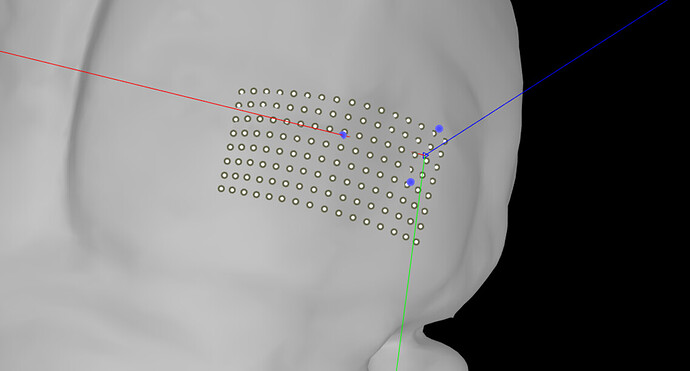
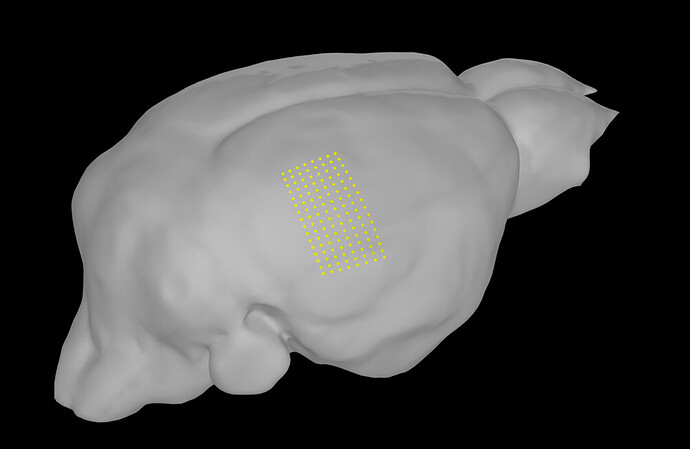
I have been using brainstorm to position a micro-ecog grid on a mouse brain, and so far it has worked great. I created a channel file with the electrode positions relative to each other (the grid has 128 channels, with spacings of 200µm in one direction and 250µm in the other direction). I then use the MRI registration function to correctly place this grid with respect to some fiducial markers.
However, I wanted to make sure the spacing between contacts is conserved when projecting the grid to the surface, as it looks distorted on the 3D volume. Is this just a visualization issue or will the projection deform the grid, and if so, how would I fix this?
Thanks!
The projection to a surface (either cortex or inner skull) will slightly change the locations as each contact is moved to the point on the surface that is closest to the current location. To check the updated distances, you could export to Matlab the Channel file and compute the distances between adjacent contacts using their locations (in ChannelFile.Channel(iChannel).Loc given in meters using SCS):
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Edit_the_channel_file
I had indeed checked this and it showed some deformation in the electrode spacing. Is there a way in which I could warp the grid to the surface in a more rigid way?
For the moment this rigid projection is not available in Brainstorm, although it is in our future developments to implement this "Folding" of grids on the cortical surface as described in:
Localization of dense intracranial electrode arrays using magnetic resonance imaging - PMC