Hello All,
When I load this file (https://www.transfernow.net/01joy1o0050a) in Brainstorm, I am getting this message below:
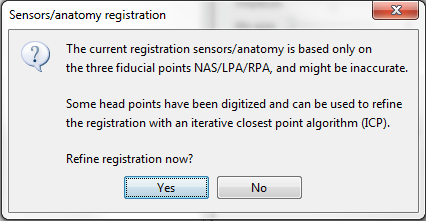
I am not familiar with electrode registration and this worries me as there might be something wrong with the electrode positions which I cannot identify. I am not sure what this message means especially as regards to digitisation and fiducials. The electrodes were imported originally into EEGLAB using standard 10/10 electrode positions and that is all.
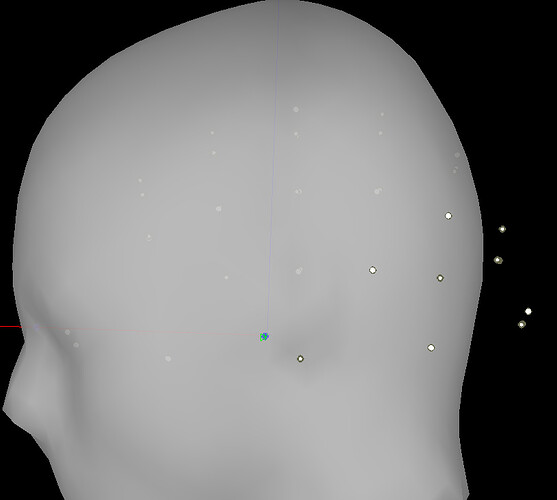
And what exactly does the message ('Refine registration now?') wants me to do. Should I use the 3D display (shown below) of electrode positions to realign the electrodes if they seem not to be in their correct positions as per the labels? When I click yes the electrodes seem to be adjusted automatically although I am not sure if I need to do anything after that. I also just do not understand why the original standard electrode positions loaded in EEGLAB might be inaccurate in Brainstorm.
I am interested in doing source localisation over these electrodes; I hope I will not have any issue as long as the electrode look visually in the right positions in Brainstorm 3D electrode display?? I just do not want source localisation results to be affected by electrode positioning problem which I am unaware.
Thanks for any clarifications. Sorry if these are very basic questions.
Bethel