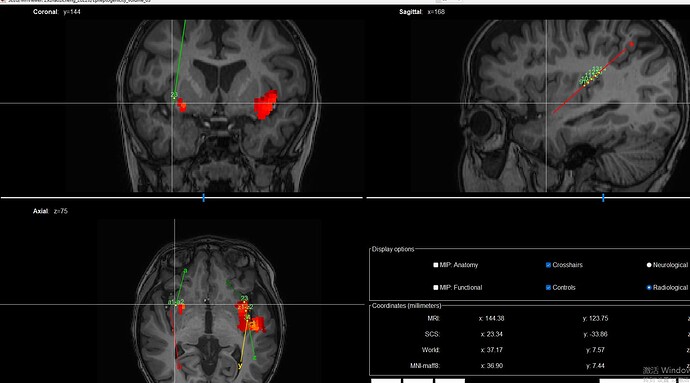
Hi, I encountered an issue while interpolating the t-values onto the anatomical images. The interpolation of the t-map is misaligned with the corresponding contact points. For example, the a-electrode that I am pointing to with the mouse does not match the corresponding t-map. Why is this happening?
Can you explain the steps you are following to perform this interpolation?
The problem seems to be solved if I select 'segment' to compute non-linear transformation between the subject space and the MNI ICBM152 space instead of 'maff8' to compute linear transformation.
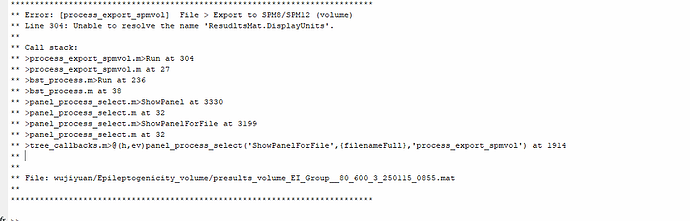
Now I encounter a new issue. There is an error when exporting the epileptogenicity volume. I want to export the T-value map as a .nii file for subsequent group study. Is there another way to do this?
This issue is solved now [Fix typo in process_export_spmvol by chinmaychinara91 · Pull Request #761 · brainstorm-tools/brainstorm3 · GitHub],
there was a typo in the code, please update your brainstorm version and try again
Thank you very much for your help. The Brainstorm is now running properly. I have another question: Is the exported t-value map in the standard MNI space in .nii format? If I want to obtain the average t-value map from multiple patients, can I simply average each patient's t-value map without needing to register them to the MNI space?
Great to hear it is working.
Regarding the group analysis, you can check this tuto:
https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Group_analysis:_MEG.2FEEG