Hello,
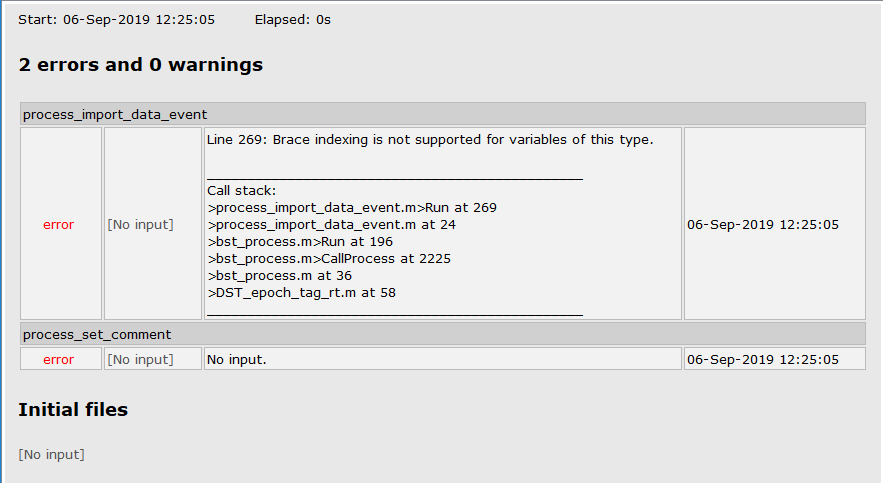
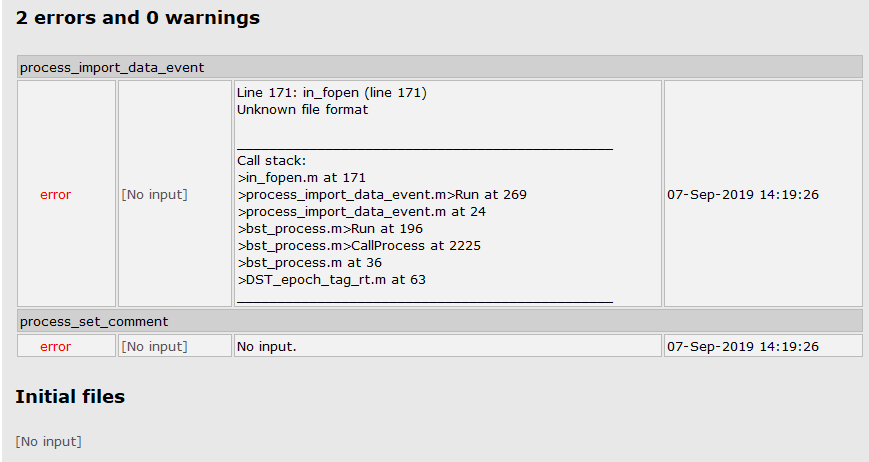
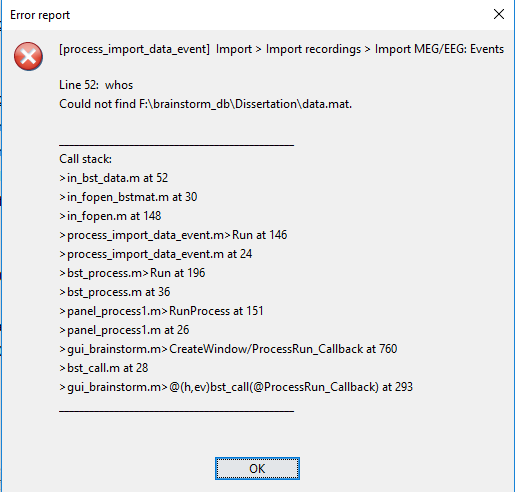
I am trying to epoch EEG raw data using my script and keep getting the following error message. I suspect this is related to the 'datafile' option, but can't figure out why. Can you help?
% Script generated by Brainstorm (25-Jul-2019)
cd 'G:\My Drive\EEG\Dissertation\Analaysis_Script\Data_Analysis'
load('Subj_list.mat');
% Input files
sFiles = ;
SubjectNames = Subj_Names;
% Create RawFiles
RawFiles1 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{1});
RawFiles2 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{2});
RawFiles3 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{3});
RawFiles4 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{4});
RawFiles5 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{5});
RawFiles6 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{6});
RawFiles7 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{7});
RawFiles8 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{8});
RawFiles9 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{9});
RawFiles10 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{10});
RawFiles11 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{11});
RawFiles12 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{12});
RawFiles13 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{13});
RawFiles14 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{14});
RawFiles15 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{15});
RawFiles16 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{16});
RawFiles17 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{17});
RawFiles18 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{18});
RawFiles19 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{19});
RawFiles20 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{20});
RawFiles21 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{21});
RawFiles22 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{22});
RawFiles23 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{23});
RawFiles24 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{24});
RawFiles25 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{25});
RawFiles26 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{26});
RawFiles27 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{27});
RawFiles28 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{28});
RawFiles29 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{29});
RawFiles30 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{30});
RawFiles31 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{31});
RawFiles32 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{32});
RawFiles33 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{33});
RawFiles34 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{34});
RawFiles35 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{35});
RawFiles36 = sprintf('F:\Dissertation\Clean_v4\%s_Cleaned_v4.set',SubjectNames{36});
RawFiles = {RawFiles1,RawFiles2,RawFiles3,RawFiles4,RawFiles5,RawFiles6,RawFiles7,RawFiles8,RawFiles9,RawFiles10,...
RawFiles11,RawFiles12,RawFiles13,RawFiles14,RawFiles15,RawFiles16,RawFiles17,RawFiles18,RawFiles19,RawFiles20,...
RawFiles21,RawFiles22,RawFiles23,RawFiles24,RawFiles25,RawFiles26,RawFiles27,RawFiles28,RawFiles29,RawFiles30,...
RawFiles31,RawFiles32,RawFiles33,RawFiles34,RawFiles35,RawFiles36};
for i = 1:1
% for i = 1:length(SubjectNames)
% Start a new report
bst_report('Start', sFiles);
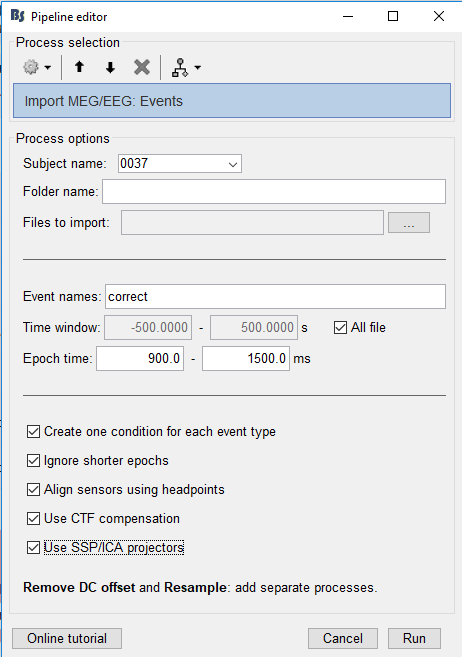
% Process: Import MEG/EEG: Events
sFiles = bst_process('CallProcess', 'process_import_data_event', sFiles, , ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'datafile', {RawFiles{1},'SET'}, ...
'eventname', 'correct', ...
'timewindow', , ...
'epochtime', [0.9, 1.5], ...
'createcond', 1, ...
'ignoreshort', 1, ...
'channelalign', 1, ...
'usectfcomp', 1, ...
'usessp', 1, ...
'freq', , ...
'baseline', [0.9, 1]);
% Process: Set comment: CRT_900-1500ms
sFiles = bst_process('CallProcess', 'process_set_comment', sFiles, [], ...
'tag', 'CRT_900-1500ms', ...
'isindex', 1);
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
%bst_report('Export', ReportFile, ExportDir);
end