Hello,
Right now, I can't display any cortical activity. For example, if I want to display the results of a source reconstruction, I get the following error message:
** Error: Line 39: Field assignment to a non-structure array object.
**
** Call stack:
** >struct_copy_fields.m at 39
** >bst_get.m>FillMissingFields at 3783
** >bst_get.m at 2892
** >panel_surface.m>SetSurfaceData at 1239
** >panel_surface.m at 38
** >view_surface_data.m at 293
** >tree_callbacks.m at 272
** >bst_call.m at 28
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 109
**
The error occurs when comparing sDest and sSrc. In this case sDest is not a structure but a char with the entry: 'tess_cortex_pial_low_keep.mat'
I can display the time series, covariances or correlation maps without any problems.
This error persisted even after the update. In addition, it came from one day to the next.
Does anyone have a suggested solution?
Thank you very much and many greetings,
Matthias
The error you report here does not correspond to the current Brainstorm version.
Please update Brainstorm and post the updated error message.
I guess the problem arises also if you display the surface itself (from the anatomy folder).
The easiest for us to debug this problem would be for you to share your subject anatomy:
- right-click on the subject > File > Duplicate subject
- delete all the files you see in the duplicated subject in the "Functional data" view, keep only the subject anatomy
- right-click on the subject > File > Export subject
- Upload the .zip somewhere (dropbox, google drive, onedrive...)
- Copy-paste the download link here
At first thank you very much for the response.
When I use the Brainstorm version from 13.10.2019 the error still occurs.
The error occurs for every subject, also when I project the data to a default brain.
But I have no problem, if I want to display the surface itself.
I uploaded the anatomy folder of one subject. Please find the link below:
https://uni-duesseldorf.sciebo.de/s/7kgfKldqP9lmFwU
The instructions I gave you were in case the error was occurring on the anatomy alone, but it doesn't. I will need some source maps in order to reproduce the error.
Please repeat the same steps, but when deleting the files, keep one source file with which you have this error, and the associated files (channel file, noise covariance, head model, recordings).
Then send me only these files: zip the folder "Protocol/data/Subject03_copy" and upload it as well.
Thanks
Please find the files with the link below.
https://uni-duesseldorf.sciebo.de/s/xXGehAqUPQwx2fZ
Let me know, if you need something else.
8.6Gb? I'm sorry, I won't be able to download this anytime soon, I work on 4G cell phone connection at the moment.
How come your files are so big? Have you kept only one source file?
Have you by mistake selected the high-resolution cortex in your anatomy folder?
If you can't package something smaller (<100 Mb), then maybe we can do this differently: post a screen capture of your database explorer showing the files that cause the problem, and copy-paste the file contents of the source file and head model file as shown by the popup menu File > View file contents.
The vectorview file was corrupt and the source file itself was big.
I was able to reduce the size to 155 Mb.
https://uni-duesseldorf.sciebo.de/s/23xctCpI4aw7dRT
At first I have the screen capture from the database and in the following i copy-paste the file content:
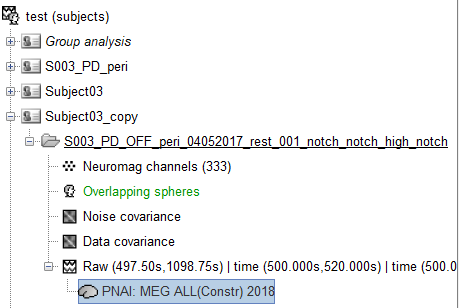
File content source file:
file_content_source_file.txt (2.8 KB)
File content head-model:
file_content_head_model.txt (37.1 KB)
Ok, thanks, I could test it.
There is nothing wrong with your database, the problem seems to be with your Brainstorm user preferences... The property DefaultSurfaceDisplay was apparently set to a string instead of a structure. I checked all the code but could not understand how this could have happened. Have you tried using bst_set() manually?
To fix the current problem, execute the following lines in your Matlab command window after starting Brainstorm:
global GlobalData;
GlobalData.Preferences = rmfield(GlobalData.Preferences, 'DefaultSurfaceDisplay');
Atlernative: type "brainstorm reset" and restart Brainstorm
Thanks a lot. Now everything is back to normal.
Yes, in one custom made script I use bst_set() manually.
I use it to set the default cortex file.
