Hello,
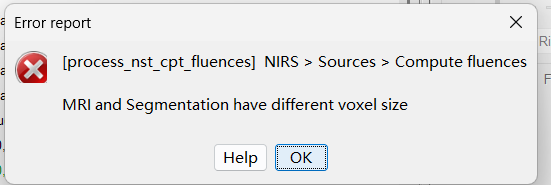
When I was trying to run the compute fluences in Nirstorm, an error popped out, saying '‘MRI and segmentation have different voxel size’.
The MRI that I used was the subject’s t1 image, and I ran the segmentation using CAT12 plugin in brainstorm:
I checked the voxel sizes of MRI and segmentation, they are the same. I dont know what caused this problem. Hope someone of you can help.
Thanks
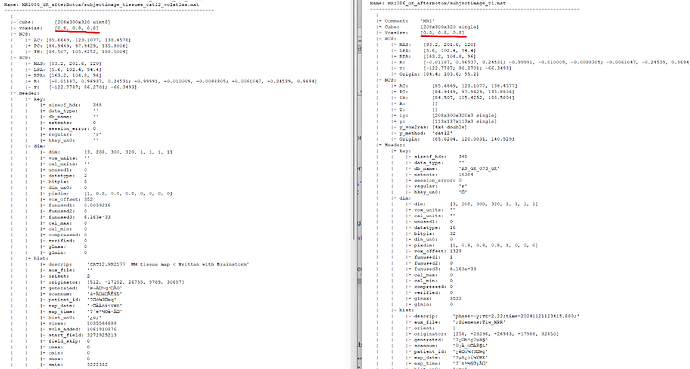
I just checked the voxel sizes of MRI and segmentation in the brainstorm dataset, and find that CAT12 segmentation slightly chaged the voxel size from 0.800000011920929 * 0.800000011920929 * 0.800000011920929 to 0.800000011920929 * 0.799999952316284 * 0.800000011920929
So how to prevent this problem?
Hello,
Thank you for the report; indeed, the dimensional difference is the issue.
@Raymundo.Cassani should brainstorm and apply some rounding on the dimensions of the MRI?
@woshibeauty one way to solve this issue would be to resample your MRI before running the segmentation; you can resample to 1x1x1 to avoid any rounding issue.
@woshibeauty, would it be possible to you to share the files?
This to determine where is the voxel size change happened, and the best way to address it.
- Duplicate such Subject
- Remove all other anatomical parcellations (to make a smaller file)
- Export the Subject
- Upload the file somewhere and post the download link either in this post of send it by direct message.
@edelaire, in places where we check for same voxel size, we do it up to 1/1000 of mm. You may want to update the check in NIRSTORM to do the same:
sameSize = all(round(sMriRef.Voxsize(1:3) .* 1000) == round(sMriSrc.Voxsize(1:3) .* 1000))
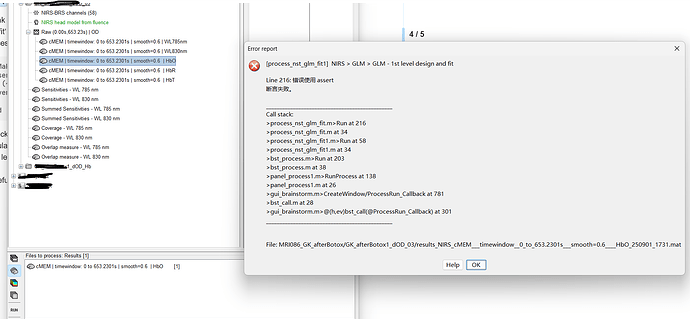
Thank you! It works well now, but I found another bug when I was trying to run ‘GLM 1st level design and fit’ based on my cMEM 3D reconstruted signal. An error was reported at line 216 of ‘process_nst_glm_fit.m‘
% Make sure time axis is consistent
assert(all(channel_data.Time == DataMat.Time));
if (~isfield(DataMat,'Events') || isempty(DataMat.Events) )&& isfield(channel_data, 'Events')
DataMat.Events = channel_data.Events;
end
I checked the time vectors in the ‘data_OD.mat’ and the ‘results_NIRS_cMEM_HbO.mat’ which was calculated based on the former one, they are not totally the same: several time points were different at e-16 level.
I think they should be the same. I don’t know which step caused these differences.
Hopefully you can help fix the code or prevent these differences.
I tried to dupicate the subject to share the files with you, but everytime something went wrong. I will try later. Thx.
Hello,
yes, when manipulating time in NIRS, we round it after the 6 digit: OPTIONS.DataTime = round(sDataIn.Time,6); ( nirstorm/bst_plugin/inverse/process_nst_cmem.m at master · Nirstorm/nirstorm · GitHub )
I need to do some checks to verify wether this is nessecary or not.
What is your sampling rate ? if the sampling rate is a nice round number, this rounding shouldnt have any effect.
any idea @Raymundo.Cassani ?
Re,
i pushed a fix here: [bugfix] GLM - better check for time consistency by Edouard2laire · Pull Request #284 · Nirstorm/nirstorm · GitHub . You should now be able to use the GLM as desired.
Please note, however, that we didn’t test using the GLM after cMEM. Notably, cMEM might distort the signal near 0 as it will try to shut down the parcel. I am not sure how those discontinuities might affect the GLM (and it was not tested).
I am currently working on an improvement of cMEM to improve on that. The preprint should be out soon; we could discuss it if you'd like.
Lastly, thanks a lot for repporting those issue to us. it is very crutial for the improvement of the software 
@edelaire
Thank you for your kind reminder.
The sampling rate is around 6.77 hz.
If cMEM has not been tested in GLM, can I use MNE instead? It will be nice if I can get the opportunity to discuss with you.
If you really need the rounding, it would be better to add it very early in the pipeline (e.g., when the raw data is linked for review).
Otherwise, you can updated the assert to consider the error margin
Hi @edelaire ,
I updated NIRSTROM, GLM works well now.
I also realize that you fixed something else: [bugfix] headmodel -- better detected old head model by Edouard2laire · Pull Request #283 · Nirstorm/nirstorm · GitHub
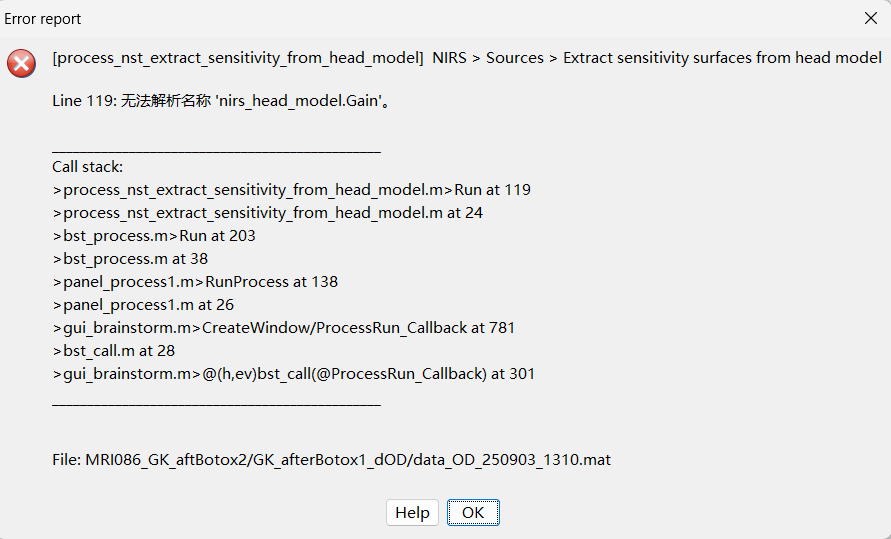
I am not sure if this modification is the reason, when I reran the ‘extract sensitivity surfaces from head model’, new error was reported
I think this line ‘if ndims(nirs_head_model.Gain) == 3‘ should be corrected to ‘if ndims(head_model.Gain) == 3‘
1 Like
Thanks a lot. it is now fixed ! i shouldn’t have merged the changes without testing