It seems that your source space is outside of the FEM Mesh.
Are you using the cortical surface as a source or the volume grid?
Ok, I'll check that and will let you know asap
@nikiita013, it seems that the cap you are using the head model are not aligned
While using the templates, you need to check visually the models.
Also, why do you want to run the 12-layer model?
You can start with simpler model, and then increase the complexity, here is the first tuto:
https://neuroimage.usc.edu/brainstorm/Tutorials/Duneuro
then you can check the advanced one with more layers.
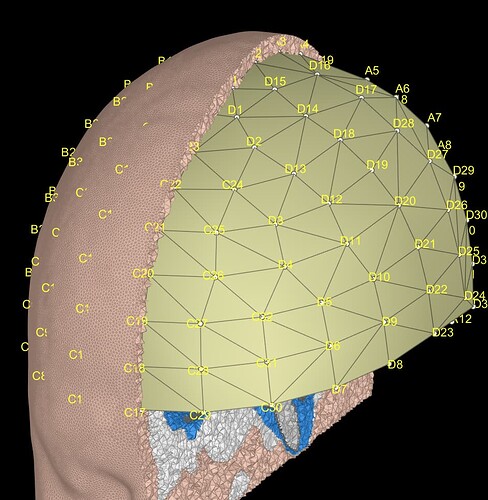
Have the cortex_15002_warped selected and 'Cortex surface' for source space when generating the DUNEuro forward modal (as I'm mostly interested in the hippocampus region).
I also made sure to tick 'Force source space inside layer "gray"
what do you mean here,
can you share a screenshot of the anatomy tab of your brainstrom?
Hi @tmedani, thanks a lot for your reply! And very sorry for late reply. I missed notifications.
This plot looks very informative. I tried to do th same plot but could not figure it. Can you please tell how you get it? Also I am actually comparing different FEM models so that is why I want to try all different variations.
Thanks!
Hi @nikiita013
You can first plot the channels
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Display_the_sensors
Then you can add the surfaces as explained here:
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Surfaces
you can use the + or - to add or remove a surface,
Let me know if you have any other questions.
Takfarinas
Hi, @tmedani
I think this case is very similar to mine.
I am still in trouble with error:'coordinate is outside of the grid,', and mergemesh/mergesurf doesn't work at my machine.
Is there anything I can do to solve this problem?
This is the past question : Encountering FEM Mesh Generation Issues with SimNIBS 4: Skull penetrate Scalp Layer - #3 by bong516612
and I can share you 2 participants who has same problem
If this is due to bad quality of MRI, than I will exclude these 2 participant from my project.
I will waiting for your opinion!
Thanks a lot,
Bong
Here is data : 2 participant's zip file (exported subject) for brainstorm, MRI (freesurfer output and T2), preprocessed .set file (EEGLAB), .pom file for channel position
https://drive.google.com/drive/folders/1CDespsqgtJ53qhDI1gXoysoHTJ9rECLL?usp=sharing
I ran 'compute head model' at the functional tab,
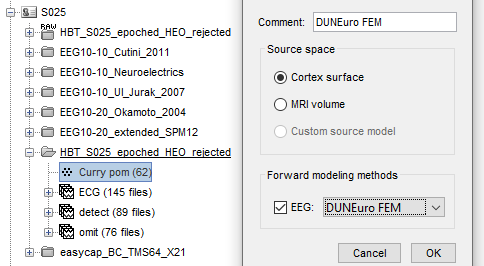
with the default option with 'Force source space inside layer "gray"' and 'Save transfer matrix'
This made 'coordinate is outside of the grid, or grid is not convex' error.
...
dipole_21875.density = sparse
dipole_21875.time = 0
dipole_21876.density = sparse
dipole_21876.time = 0
dipole_21877.density = sparse
dipole_21877.time = 0
dipole_21878.density = sparse
dipole_21878.time = 0
dipole_21879.density = sparse
dipole_21879.time = 0
dipole_21880.density = sparse
dipole_21880.time = 0
dipole_21881.density = sparse
dipole_21881.time = 0
dipole_21882.density = sparse
dipole_21882.time = 0
dipole_21883.density = sparse
dipole_21883.time = 0
dipole_21884.density = sparse
dipole_21884.time = 0.0009983
dipole_21885.density = sparse
dipole_21885.time = 0
dipole_21886.density = sparse
dipole_21886.time = 0
dipole_21887.density = sparse
dipole_21887.time = 0
dipole_21888.density = sparse
dipole_21888.time = 0
dipole_21889.density = sparse
dipole_21889.time = 0
dipole_21890.density = sparse
dipole_21890.time = 0
dipole_21891.density = sparse
dipole_21891.time = 0
dipole_21892.density = sparse
dipole_21892.time = 0
dipole_21893.density = sparse
dipole_21893.time = 0
dipole_21894.density = sparse
dipole_21894.time = 0
dipole_21895.density = sparse
dipole_21895.time = 0
dipole_21896.density = sparse
dipole_21896.time = 0
dipole_21897.density = sparse
dipole_21897.time = 0
dipole_21898.density = sparse
dipole_21898.time = 0
dipole_21899.density = sparse
dipole_21899.time = 0
dipole_21900.density = sparse
dipole_21900.time = 0
dipole_21901.density = sparse
dipole_21901.time = 0
dipole_21902.density = sparse
dipole_21902.time = 0
dipole_21903.density = sparse
dipole_21903.time = 0
dipole_21904.density = sparse
dipole_21904.time = 0.0010035
dipole_21905.density = sparse
dipole_21905.time = 0
dipole_21906.density = sparse
dipole_21906.time = 0
dipole_21907.density = sparse
dipole_21907.time = 0
dipole_21908.density = sparse
dipole_21908.time = 0
dipole_21909.density = sparse
dipole_21909.time = 0
dipole_21910.density = sparse
dipole_21910.time = 0
dipole_21911.density = sparse
dipole_21911.time = 0
dipole_21912.density = sparse
dipole_21912.time = 0
dipole_21913.density = sparse
dipole_21913.time = 0
dipole_21914.density = sparse
dipole_21914.time = 0
dipole_21915.density = sparse
dipole_21915.time = 0.0010009
dipole_21916.density = sparse
dipole_21916.time = 0
dipole_21917.density = sparse
dipole_21917.time = 0
dipole_21918.density = sparse
dipole_21918.time = 0
dipole_21919.density = sparse
dipole_21919.time = 0
dipole_21920.density = sparse
dipole_21920.time = 0
dipole_21921.density = sparse
dipole_21921.time = 0
dipole_21922.density = sparse
dipole_21922.time = 0
dipole_21923.density = sparse
dipole_21923.time = 0
dipole_21924.density = sparse
dipole_21924.time = 0
dipole_21925.density = sparse
dipole_21925.time = 0
dipole_21926.density = sparse
dipole_21926.time = 0
dipole_21927.density = sparse
dipole_21927.time = 0
dipole_21928.density = sparse
dipole_21928.time = 0
dipole_21929.density = sparse
dipole_21929.time = 0
dipole_21930.density = sparse
dipole_21930.time = 0
dipole_21931.density = sparse
dipole_21931.time = 0
dipole_21932.density = sparse
dipole_21932.time = 0
dipole_21933.density = sparse
dipole_21933.time = 0
dipole_21934.density = sparse
dipole_21934.time = 0.0010014
dipole_21935.density = sparse
dipole_21935.time = 0
dipole_21936.density = sparse
dipole_21936.time = 0
dipole_21937.density = sparse
dipole_21937.time = 0
dipole_21938.density = sparse
dipole_21938.time = 0
dipole_21939.density = sparse
dipole_21939.time = 0
dipole_21940.density = sparse
dipole_21940.time = 0
dipole_21941.density = sparse
dipole_21941.time = 0
dipole_21942.density = sparse
dipole_21942.time = 0
dipole_21943.density = sparse
dipole_21943.time = 0
dipole_21944.density = sparse
dipole_21944.time = 0
dipole_21945.density = sparse
dipole_21945.time = 0
dipole_21946.density = sparse
dipole_21946.time = 0
dipole_21947.density = sparse
dipole_21947.time = 0
dipole_21948.density = sparse
dipole_21948.time = 0
dipole_21949.density = sparse
dipole_21949.time = 0
dipole_21950.density = sparse
dipole_21950.time = 0
dipole_21951.density = sparse
dipole_21951.time = 0
dipole_21952.density = sparse
dipole_21952.time = 0
dipole_21953.density = sparse
dipole_21953.time = 0
Dune reported error: Dune::Exception [findEntityImpl:/home/juan/bst-duneuro/src/duneuro/duneuro/common/edgehopping.hh:84]: coordinate is outside of the grid, or grid is not convex
status =
-1
***************************************************************************
** Error: Error during the DUNEuro computation, see logs in the command window.
***************************************************************************
>>
is there any suggestions?
Ping @tmedani
Hi @bong516612
I apologize for the delay in responding to this issue.
We have been investigating the problem and reached out to the Duneuro development team.
It appears that, in some cases, certain default parameters in the Duneuro source code are suboptimal.
The team will be making changes to the source code, and we will release a new version of the Duneuro binary once those updates are completed on their end.
In the meantime, you can try modifying the cortex file by generating a new surface (using the mesh option -right-click on the cortex file-) or manually identifying the problematic vertex—dipole 24953 in this case—and slightly adjusting its position.
Thank you @tmedani. It was a great help.
I’m looking forward to the release of the new binary.
Aside from that, using the GUI to locate a problematic vertex on the surface and then reposition it seems quite limited (I can't see how to change single vertex on display tab..).
Importing the surface (in this case, mid_15002V?) into the MATLAB Workspace to find the location of that vertex, move it, and then re-checking in the Brainstorm GUI is quite difficult.
However, referring to the answer you posted in another thread, and watching the linked video:
By comparing mid_15002V with cortex_fem and slightly reducing mid_15002V, FEM was successfully calculated on the surface.
I’m not sure if this is a general solution, but I’m posting this in case it helps anyone else experiencing a similar issue. Thank you.
Thank you, @bong516612, for the feedback!
It is great to hear that everything is working fine now.
we will work on it; it may require some time since the release may have some other features.
We will post an announcement once it is within Brainstorm.
Indeed this feature is not on Brainstorm as it is not very common to change the location of the few vertices. It requires some work to add it in the main interface, we will add it to the toDo list.
By comparing mid_15002V with cortex_fem and slightly reducing mid_15002V, FEM was successfully calculated on the surface.
I’m not sure if this is a general solution, but I’m posting this in case it helps anyone else experiencing a similar issue. Thank you.
Great to hear that the solution is working. Again, this is also related to a few dipoles.
While this is not an optimal solution, if the shift of the new surface is just by a few mm [2 to 3mm], it is still acceptable, given the high resolution of the cortex and the FEM mesh.
Thanks again for sharing your feedback, this will helpother users for sure.
Best,