Hi everyone,
I performed First-Level analysis using FreeSurfer for a certain contrast. Now I am going to load the results into BrainStorm to use the activated area as seed points; however, I couldn't find any direct way to load the sig.nii.gz file as an overlay to the cortex model in BrainStorm.
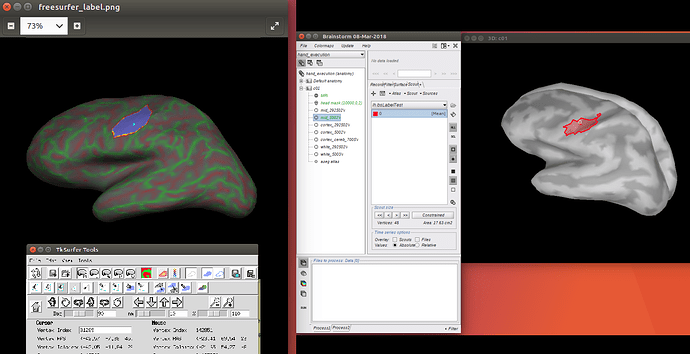
I then used tksurfer to take the activated area as ROI and create a label for that. I stored it as *.label file. Then according to what I've read in the Forum, I tried to create a new Atlas and load the label file as an atlas; however, I got an error due to the different number of vertices.
I tried that for the lh of the cortex which has 145404 vertices in my *.label file. The whole cortex however has 292502 vertices according to BrainStorm. It seems that BrainStorm tries to split the lh and rh somehow which at the end the number of vertices is not compatible with the *.label file.
I also checked the other *label files which had been created automatically by FreeSurfer in its "label" directory. The number of vertices that they have (145192) is also different from what I got in my *.label file! And they work when I read them as a new atlas (and displaying the full resolution cortex model).
I'm totally confused how everything works with the veritces and how BrainStorm reads the anatomy from the FreeSurfer's folder.
Is there any direct way reading FreeSurfer's sig.nii.gz or tksconcat.nii files as an overlay in BrainStorm?
Could you please help me with that?
Kind regards
Tannaz