Hi Brainstorm Team,
I'm trying to perform unsupervised spike sorting with wave_clus and ultramegasort2000.
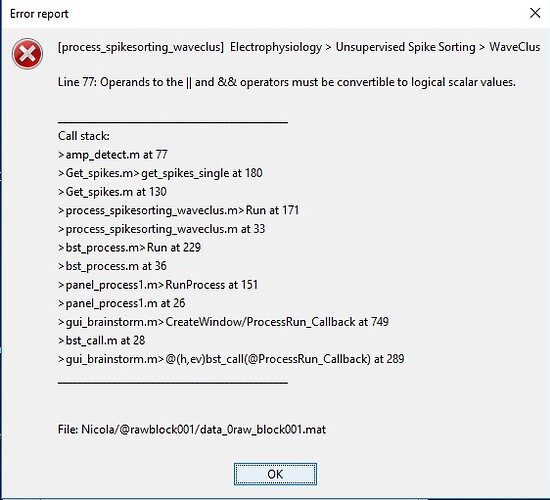
This is the same error report for different .edf files, using wave_clus:
The report appears just at the beginning of extraction spikes process.
Following instead report error performing Ultramegasort2000:
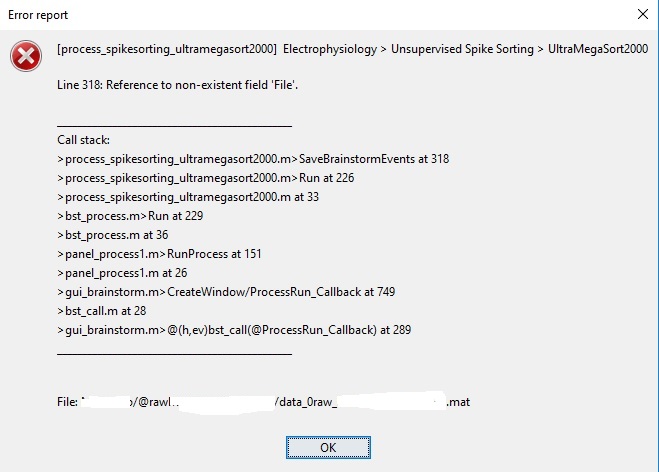
At the contrary the report appears at the end of extraction spikes process.
Any idea what could be the problem?
Thank you in advance,
Matteo
Hi Matteo,
for the second error, this would occur if no files were created during the spike-sorting process;
this means that either you have low sampling rate on your raw data, or there were absolutely no spikes on ALL your electrodes (highly unlikely). Try to lower the threshold on the parameters option on the main UltraMegaSort2000 window: spikes.params.thresh
The first error seems to be inside the WaveClus spike-sorter and I've never came across this:
can you send a link to download one example .edf file that you used to take a look?
Thanks,
Konstantinos
1 Like
Hi Konstantinos,
first of all thanks for your kind reply.
I sent to you a link to download one example .edf file by private message.
I can detect spikes visually with higher amplitude in C3/P3 electrodes.
Thank you again in advance.
Matteo
Hi @mcataldi,
I just opened your file and unless there is something wrong, it seems to be sampled at 128 Hz.
I assume your dataset is from an EEG recording (since you mentioned C3/P3 electrodes) sampled at 128 Hz, or downsampled at 128 Hz.
The toolbox that does spike-sorting is made for Invasive Neurophysiology which takes raw high-sampled data from invasive recordings (Fs>20,0000 Hz).
The notation of a "spike" in this toolbox doesn't refer to something that looks like a spike, it literally refers to spikes that are picked up from individual neurons.
You won't be able to use this toolbox for your purposes.
However you should be able to use other tools to cover your needs through Brainstorm.
Hope that helps,
Konstantinos