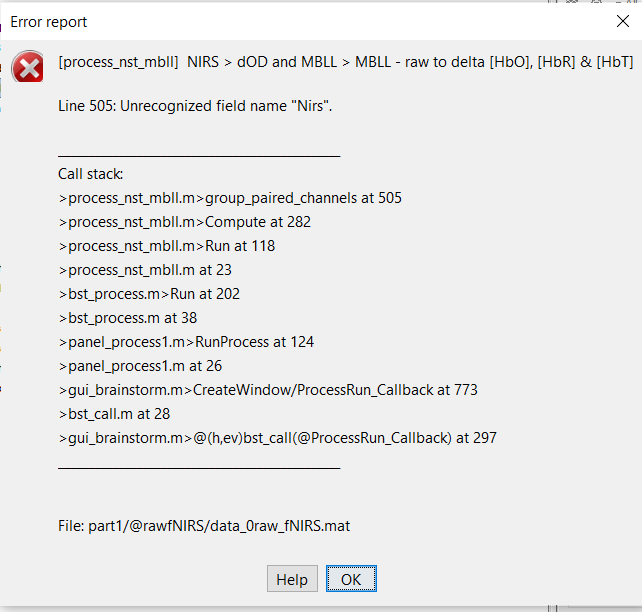
I collected 16 channels of fNIRS data from 8 sensors placed on an EEG cap and put them into a matrix of channels x time. This is a .mat file. I'm trying to see if I can follow the NIRS finger tapping tutorial and use NIRSTORM for analysis, but every time I try and do the MBLL - raw to delta [HbO], [HbR] & [HbT] process I get the error above.
I have no idea what this error is, and I don't know what it is referring to. Do I need something added to my code? I also have problems uploading my data to brainstorm because there isn't a ".mat" option that doesn't use Volts as the amplitude. Is there a script to turn the .mat to a .nirs?
My brainstorm version is Version: 3.220525 and my NIRSTORM version is 1.6.91.
Thank you! I'm sorry if I didn't explain anything well enough, this is my first time analyzing fNIRS.
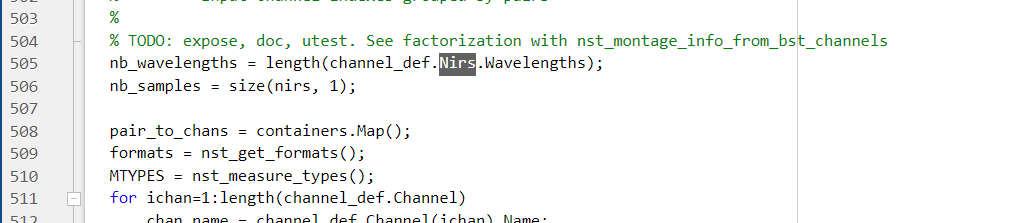
The code snippet from the MBLL process is as follows:
Hello,
Could you clarify how you imported the NIRS data inside brainstorm ? Currently, if you imported data in another format than .nirs or .snirf; some field essential for NIRSTORM might not have been set properly.
If you share one of your files here; we might be able to help change the current data importation; but i think the easiest would be to convert your data into a format supported by Nirstorm first (.nirs - easiest i think or .snirf). You can check FAQ | HOMER2 about the .nirs format
Edouard
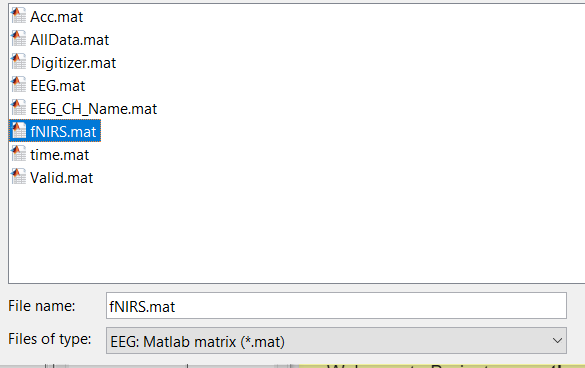
I imported it as an eeg matrix .mat file. It only has the data x time for the 8 sensors in the file, so I am missing location data. It looks like there are a bunch of fields I don't have data for, do you think it would be possible to make a .nirs formatted file with just the data I have?