Hello everyone,
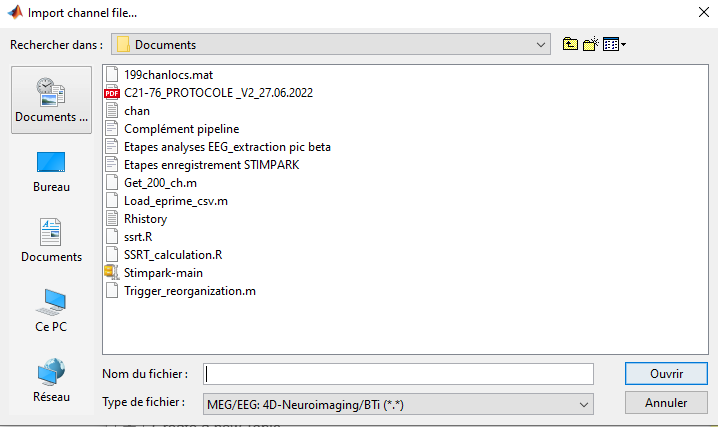
I have trouble importing a .mat channels file. When I use the Import channel file from right-clicking on the subject, the file I want to use does not appear when I select the correct file format (.mat). Trying to bypass this by selecting another file format (my .mat file then appears in the window) leads to an error message.
Error: Line 50: in_fopen_4d (line 50)
** Error reading 4D/Bti recordings: No config file.
** If you are trying to read recordings from another acquisition system,
** please select the appropriate file format in the "Open file" dialog window.
**
** Call stack:
** >in_fopen_4d.m at 50
** >in_fopen.m at 77
** >import_channel.m at 118
** >tree_callbacks.m>ImportChannelCheck at 3644
** >bst_call.m at 28
** >tree_callbacks.m>@(h,ev)bst_call(@ImportChannelCheck,iAllStudies) at 2762
I am currently using Version: 3.220131 (31-Jan-2022) on R2020b, win64.
Thanks in advance for your help!
Sina
And here is what happens when I choose the right file format.
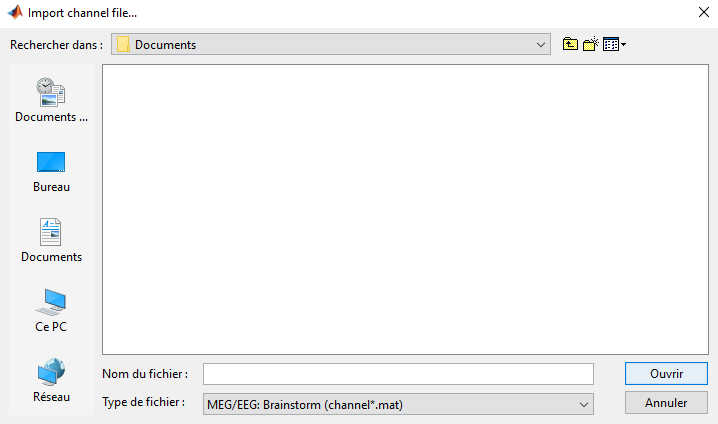
Looks like the file name needs to start with 'channel'. Rename your file to channel199chanlocs.mat and please give it another try.
Hi Sina,
Keep in mind that .mat files are not a file format, but rather they are containers for Matlab variables.
Thus data can be organized in many different ways inside a .mat file.
The format MEG/EEG: Brainstorm (channel*.mat) is expecting a Brainstorm channel file (https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#On_the_hard_drive). As Sylvain mentioned, these files usually have the word channel at the beginning.
- How was your
.mat file created?
- On Matlab? With other software?
- How is its content organized?
Hi Sylvain, Hi Raymundo,
Many thanks for your help, indeed, renaming the file allows it to be selected. However, I get this error message in the command window.
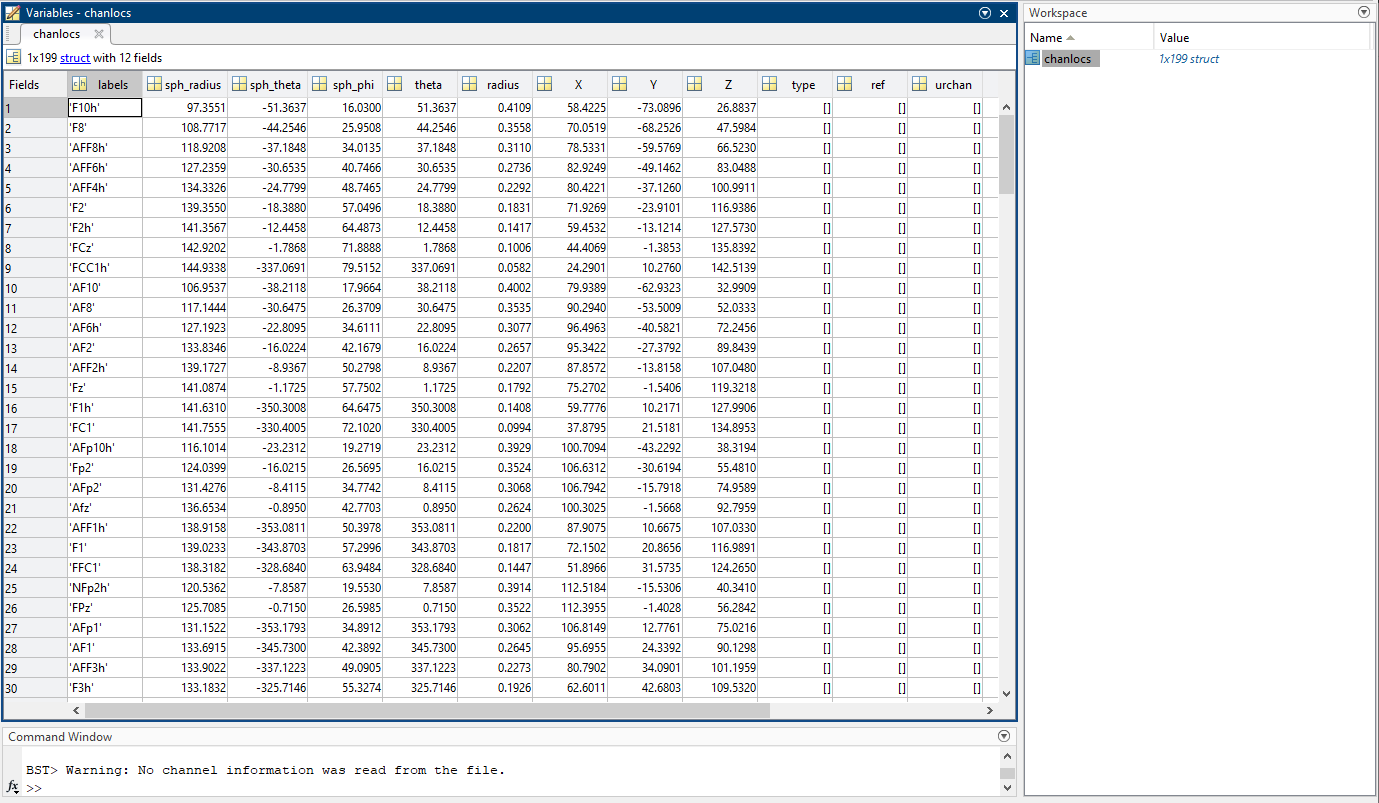
This is the file my colleagues use for our protocol, I am afraid that I do not have many informations on how it was created. The screenshot gives an idea about its content.
Sina